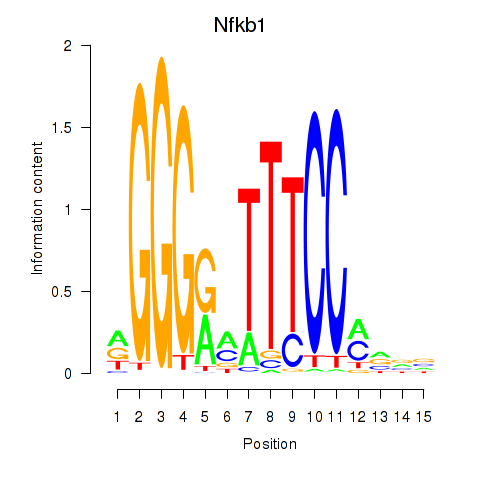
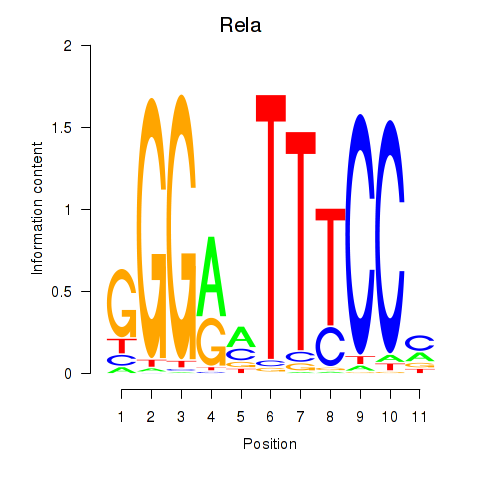
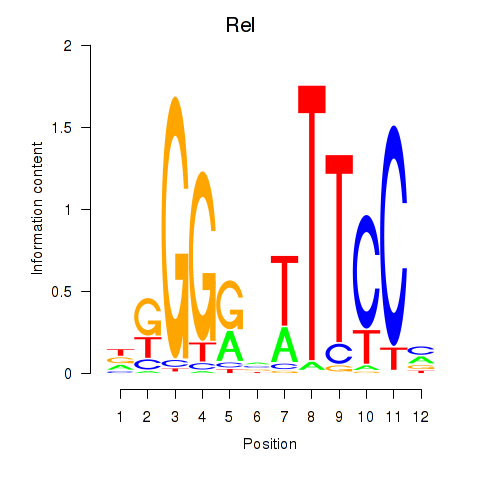
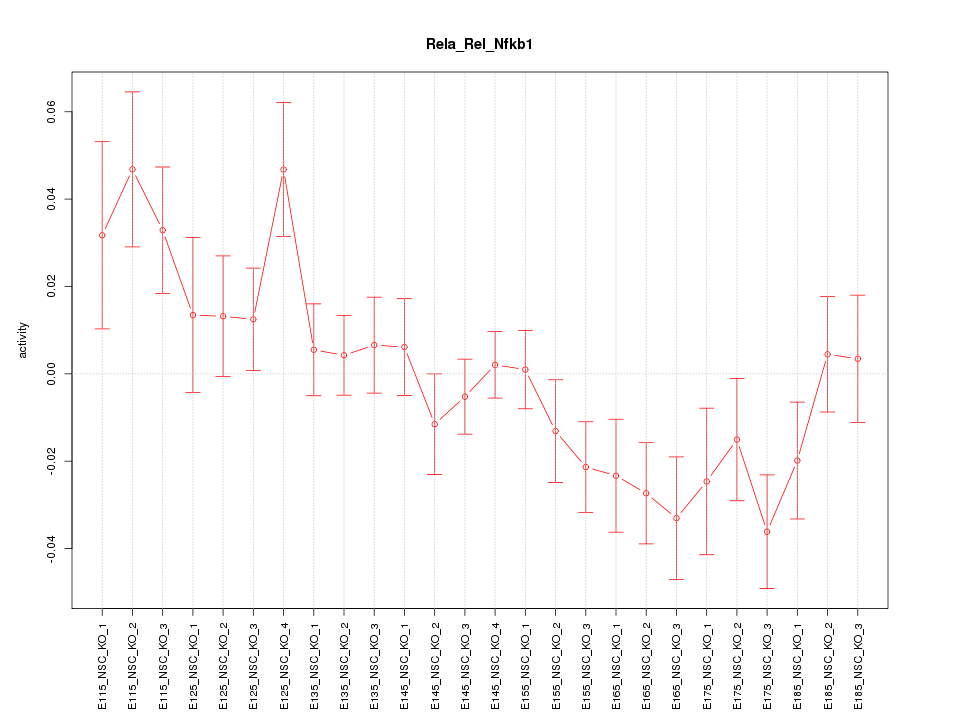
| 2.1 |
10.4 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 1.5 |
4.6 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 1.4 |
10.1 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.3 |
4.0 |
GO:0032916 |
positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 1.0 |
6.2 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 1.0 |
3.9 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 1.0 |
3.8 |
GO:1903416 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 0.9 |
4.5 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.9 |
3.5 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.9 |
8.5 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.8 |
2.5 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.8 |
5.6 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.8 |
2.4 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) primary alcohol catabolic process(GO:0034310) |
| 0.8 |
4.6 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.7 |
2.1 |
GO:0070103 |
regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.7 |
2.8 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.7 |
6.7 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.6 |
1.9 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.6 |
6.7 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.6 |
2.4 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 |
4.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.6 |
3.4 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.6 |
1.7 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.6 |
1.7 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.5 |
0.5 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.5 |
9.7 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.5 |
1.5 |
GO:0090403 |
regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of fermentation(GO:0043465) oxidative stress-induced premature senescence(GO:0090403) negative regulation of fermentation(GO:1901003) |
| 0.5 |
1.5 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.5 |
2.4 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) negative regulation of filopodium assembly(GO:0051490) |
| 0.4 |
3.5 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.4 |
5.5 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.4 |
1.2 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 |
3.1 |
GO:0006490 |
oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.4 |
1.2 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.4 |
2.7 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.4 |
1.5 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.4 |
1.9 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.4 |
1.1 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.4 |
1.1 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.4 |
3.2 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.3 |
3.0 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.3 |
1.6 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.3 |
1.6 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.3 |
1.0 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 |
3.2 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 |
3.7 |
GO:2000254 |
regulation of male germ cell proliferation(GO:2000254) |
| 0.3 |
1.5 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 |
3.1 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.3 |
2.7 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 |
1.7 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.3 |
0.8 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.3 |
3.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 |
0.5 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.3 |
3.2 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.3 |
2.1 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.3 |
0.8 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.3 |
0.8 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 |
1.5 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.2 |
0.7 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.2 |
1.7 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 |
2.5 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 |
0.9 |
GO:0030050 |
vesicle transport along actin filament(GO:0030050) |
| 0.2 |
0.9 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 |
1.2 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.2 |
0.9 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 |
0.7 |
GO:1904884 |
telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 |
2.3 |
GO:0034433 |
steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 |
0.6 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 |
0.4 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.2 |
0.8 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.2 |
1.0 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.2 |
0.4 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 |
0.6 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.2 |
0.6 |
GO:0033030 |
negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.2 |
2.1 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.2 |
0.2 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 |
2.2 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 |
0.7 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 |
2.9 |
GO:0002467 |
germinal center formation(GO:0002467) |
| 0.2 |
0.9 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 |
0.5 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) |
| 0.2 |
0.3 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 |
3.0 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.2 |
0.8 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 |
1.1 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 |
0.5 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.2 |
0.6 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 |
0.8 |
GO:0060536 |
cartilage morphogenesis(GO:0060536) |
| 0.2 |
6.2 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 |
0.6 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 |
0.6 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 |
2.2 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
8.5 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 |
1.2 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.1 |
1.1 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 |
1.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
0.4 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.1 |
1.0 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 |
1.2 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.6 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
0.6 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.4 |
GO:1901856 |
negative regulation of cellular respiration(GO:1901856) |
| 0.1 |
0.7 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.1 |
1.0 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.1 |
4.5 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.5 |
GO:0032621 |
interleukin-18 production(GO:0032621) |
| 0.1 |
0.6 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 |
2.0 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.6 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 |
1.2 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 |
0.4 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.1 |
0.7 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.1 |
0.7 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) trophectodermal cell proliferation(GO:0001834) |
| 0.1 |
0.9 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.1 |
2.2 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.1 |
0.5 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.1 |
0.7 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 |
2.0 |
GO:1900027 |
regulation of ruffle assembly(GO:1900027) |
| 0.1 |
0.5 |
GO:0070633 |
transepithelial transport(GO:0070633) |
| 0.1 |
0.8 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 |
0.3 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 |
0.4 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 |
0.8 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.1 |
0.5 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.1 |
2.4 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.1 |
0.8 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.1 |
2.1 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
1.1 |
GO:0097066 |
response to thyroid hormone(GO:0097066) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
1.2 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 |
0.3 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
7.2 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 |
1.0 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
2.2 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.9 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.1 |
0.4 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) regulation of cholesterol import(GO:0060620) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 |
0.3 |
GO:0035964 |
vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
1.5 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.1 |
0.5 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.3 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.5 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
0.6 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 |
0.3 |
GO:0072361 |
regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 |
3.8 |
GO:0002066 |
columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 |
1.1 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 |
0.8 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
1.8 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 |
0.3 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.5 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.1 |
0.9 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.6 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.1 |
0.5 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.1 |
0.6 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 |
0.3 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.1 |
1.6 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
2.4 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.1 |
0.1 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.5 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 |
0.3 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.1 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
0.5 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
1.3 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.1 |
0.5 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 |
0.9 |
GO:0097062 |
dendritic spine maintenance(GO:0097062) |
| 0.1 |
1.9 |
GO:1901222 |
regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 |
0.5 |
GO:0070571 |
negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 |
0.7 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.1 |
1.6 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
0.2 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.1 |
1.7 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.1 |
0.2 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.5 |
GO:0006310 |
DNA recombination(GO:0006310) |
| 0.1 |
0.4 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 |
1.3 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 |
0.4 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.0 |
0.6 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.3 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.0 |
0.9 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.6 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.6 |
GO:0019884 |
antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 |
2.6 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
1.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.2 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.5 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
4.2 |
GO:2000045 |
regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 |
0.5 |
GO:0046686 |
response to cadmium ion(GO:0046686) |
| 0.0 |
0.4 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
1.7 |
GO:0035082 |
axoneme assembly(GO:0035082) |
| 0.0 |
2.3 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
0.2 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.1 |
GO:0072592 |
integrin biosynthetic process(GO:0045112) regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.0 |
0.2 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.7 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.3 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) |
| 0.0 |
0.2 |
GO:0002834 |
immune response to tumor cell(GO:0002418) regulation of response to tumor cell(GO:0002834) regulation of immune response to tumor cell(GO:0002837) |
| 0.0 |
0.8 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.2 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 |
0.7 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
1.3 |
GO:0000281 |
mitotic cytokinesis(GO:0000281) |
| 0.0 |
0.3 |
GO:0048096 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.2 |
GO:0061032 |
chorio-allantoic fusion(GO:0060710) visceral serous pericardium development(GO:0061032) |
| 0.0 |
0.5 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.1 |
GO:0072369 |
regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 |
1.1 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.1 |
GO:0031953 |
negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 |
4.2 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) |
| 0.0 |
0.6 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.9 |
GO:0048661 |
positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 |
0.8 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.9 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
1.4 |
GO:0060021 |
palate development(GO:0060021) |
| 0.0 |
0.1 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.3 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
1.3 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.2 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
1.1 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.0 |
0.1 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 |
0.6 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.9 |
GO:0002209 |
behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 |
0.6 |
GO:0000460 |
maturation of 5.8S rRNA(GO:0000460) |
| 0.0 |
0.1 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 |
0.2 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.1 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.2 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.4 |
GO:0033233 |
regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.2 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.3 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.1 |
GO:0006481 |
C-terminal protein methylation(GO:0006481) |
| 0.0 |
0.1 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 |
0.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.3 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.2 |
GO:0043954 |
cellular component maintenance(GO:0043954) |
| 0.0 |
0.1 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.5 |
GO:0042633 |
molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 |
0.5 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.1 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |