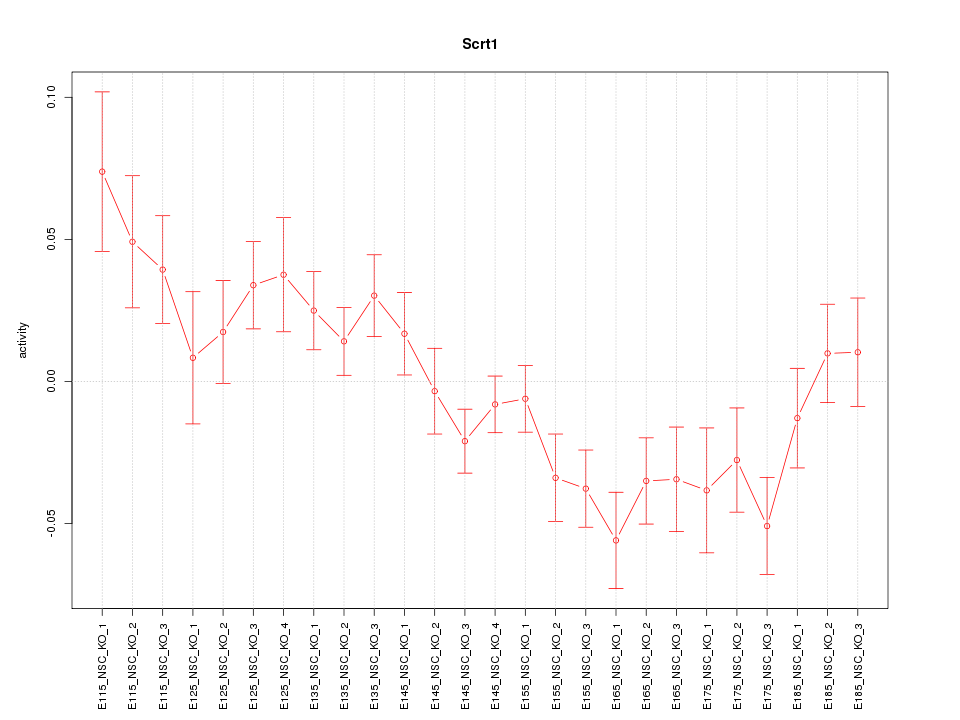
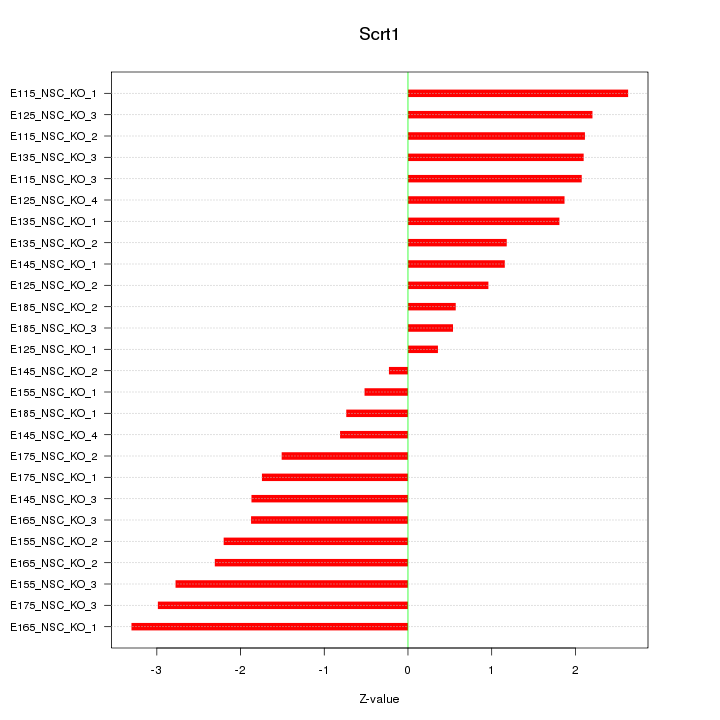
Motif ID: Scrt1
Z-value: 1.836
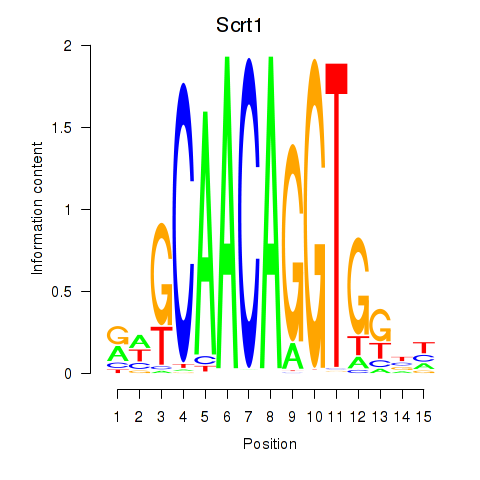
Transcription factors associated with Scrt1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Scrt1 | ENSMUSG00000048385.8 | Scrt1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt1 | mm10_v2_chr15_-_76521902_76522129 | -0.77 | 4.7e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 30.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.4 | 7.2 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 2.0 | 6.1 | GO:0086069 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) intermediate filament bundle assembly(GO:0045110) maintenance of organ identity(GO:0048496) bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 1.6 | 11.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.6 | 4.9 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 1.5 | 6.0 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 1.5 | 8.7 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 1.2 | 5.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.9 | 15.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.8 | 5.9 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.7 | 2.1 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.7 | 6.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.7 | 4.9 | GO:1901724 | positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.7 | 1.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.7 | 2.0 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.6 | 2.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.6 | 3.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.6 | 3.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.6 | 2.3 | GO:0015744 | succinate transport(GO:0015744) |
| 0.6 | 2.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.5 | 5.7 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.5 | 2.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.5 | 6.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 2.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.4 | 2.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.4 | 1.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.4 | 1.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.4 | 4.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.4 | 2.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 1.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.3 | 3.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 2.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.3 | 4.7 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 4.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 1.7 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.3 | 1.7 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.3 | 0.5 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.3 | 1.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.3 | 0.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 7.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.3 | 1.6 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.3 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 1.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 3.8 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 1.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.2 | 8.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 0.7 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 1.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 3.1 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 3.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 0.8 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.2 | 2.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 5.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 2.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 4.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 1.7 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.2 | 1.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 0.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.6 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.2 | 0.9 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 4.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 3.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.5 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.4 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 1.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 0.4 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.1 | 1.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 2.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 2.9 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 2.9 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 3.6 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.0 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 2.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 2.2 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.1 | 3.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 1.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 1.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 1.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 5.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 2.4 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.1 | 1.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.3 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 1.4 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.1 | 2.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.6 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 1.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 3.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 1.7 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 3.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 2.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.1 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.5 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 2.7 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 7.0 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.0 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 2.4 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 30.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.2 | 6.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.8 | 3.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.7 | 7.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.6 | 6.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 1.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.4 | 3.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 5.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 3.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 2.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 5.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 11.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 2.0 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 2.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.9 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) Flemming body(GO:0090543) |
| 0.1 | 1.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 4.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 6.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 9.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 7.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 2.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 3.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 3.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 3.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 4.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 15.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 4.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 6.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 5.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 3.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 31.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.7 | 5.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.6 | 7.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.4 | 7.2 | GO:0071253 | connexin binding(GO:0071253) |
| 1.3 | 3.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.2 | 4.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.2 | 3.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 4.0 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.7 | 6.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.6 | 2.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.6 | 2.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.6 | 2.3 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.5 | 2.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.5 | 6.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 1.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 2.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.5 | 11.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 2.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 8.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.4 | 3.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 4.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 2.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 5.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.4 | 5.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 4.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 5.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 0.8 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) |
| 0.2 | 6.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.2 | 2.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.0 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 2.7 | GO:0005536 | glucose binding(GO:0005536) |
| 0.2 | 3.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 13.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 1.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 0.7 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 3.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 4.7 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.1 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 4.3 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 13.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 3.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 3.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 3.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 3.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 3.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.5 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 3.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |