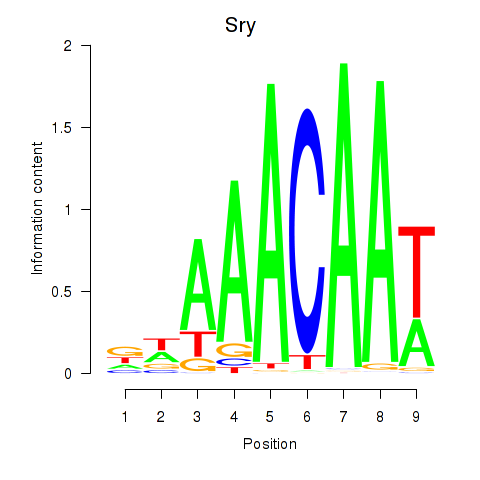
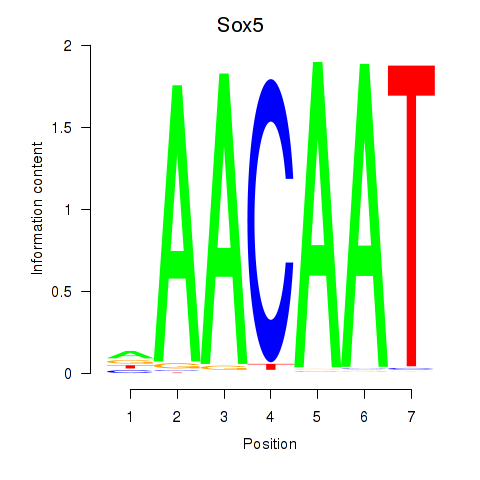
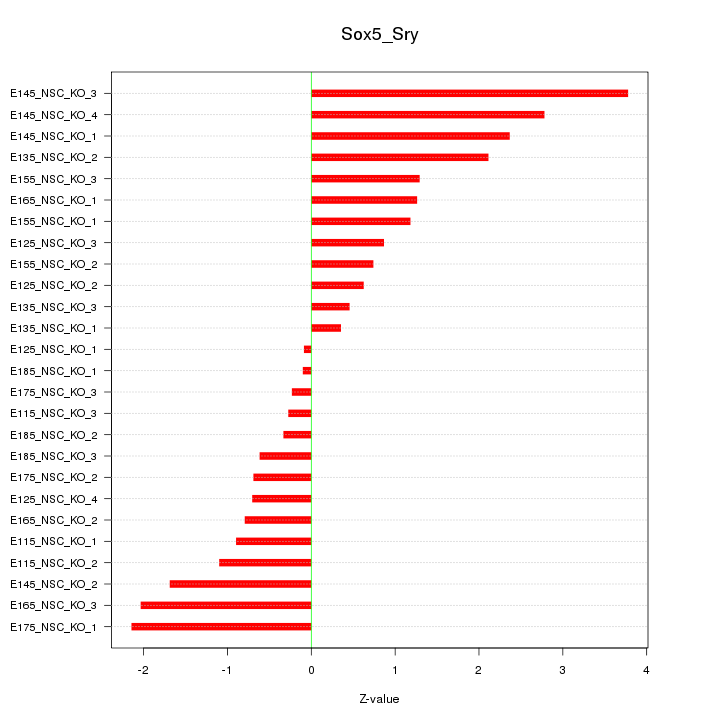
| 2.6 |
10.4 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.2 |
3.5 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.7 |
2.1 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.5 |
4.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 |
1.6 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 |
1.1 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.3 |
0.9 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 |
2.0 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 |
1.9 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.3 |
1.1 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.3 |
1.0 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.2 |
2.6 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 |
2.2 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 |
0.6 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.2 |
1.8 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.2 |
1.8 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.2 |
0.7 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.2 |
0.7 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.2 |
0.5 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 |
0.5 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.2 |
0.7 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 |
2.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.2 |
0.6 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) general adaptation syndrome(GO:0051866) |
| 0.2 |
1.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.2 |
0.2 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
1.0 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 |
0.4 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 |
0.6 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 |
0.4 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 |
0.7 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 |
0.3 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.1 |
0.4 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 |
0.8 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
1.4 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
0.5 |
GO:0061428 |
embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
1.8 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.5 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
1.0 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.1 |
1.0 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
1.0 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
0.8 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.5 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.1 |
0.5 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.2 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
0.6 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 |
0.5 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
1.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
1.2 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 |
0.7 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.5 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
0.9 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.1 |
1.0 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.7 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
0.2 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 |
0.2 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.4 |
GO:0071600 |
otic vesicle morphogenesis(GO:0071600) |
| 0.1 |
1.3 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
1.0 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.1 |
GO:0060160 |
musculoskeletal movement, spinal reflex action(GO:0050883) negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 |
0.4 |
GO:1902965 |
protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 |
3.2 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 |
0.3 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 |
0.4 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 |
0.7 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.2 |
GO:0060423 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.1 |
0.3 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 |
0.4 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 |
0.4 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.1 |
0.4 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
1.5 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 |
0.1 |
GO:1905050 |
positive regulation of metallopeptidase activity(GO:1905050) |
| 0.1 |
0.3 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 |
1.0 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.1 |
0.9 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
1.9 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
0.2 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.1 |
0.5 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 |
0.6 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) regulation of somitogenesis(GO:0014807) |
| 0.1 |
0.6 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 |
0.4 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 |
1.7 |
GO:0021799 |
cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.1 |
0.6 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 |
0.4 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.3 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.0 |
0.5 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.1 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.3 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.2 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.1 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
0.1 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 |
0.8 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
2.6 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.2 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.4 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.6 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 |
0.2 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.0 |
0.5 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.2 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.0 |
0.6 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.2 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.1 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.0 |
0.1 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.0 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.0 |
0.7 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.1 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 |
0.1 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
0.1 |
GO:1900181 |
negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 |
0.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 |
0.3 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.5 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.4 |
GO:0051443 |
positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 |
0.8 |
GO:0051017 |
actin filament bundle assembly(GO:0051017) |
| 0.0 |
0.6 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.0 |
0.5 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.3 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.2 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.1 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.2 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.4 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.3 |
GO:0016071 |
mRNA metabolic process(GO:0016071) |
| 0.0 |
0.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
1.5 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
0.1 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |