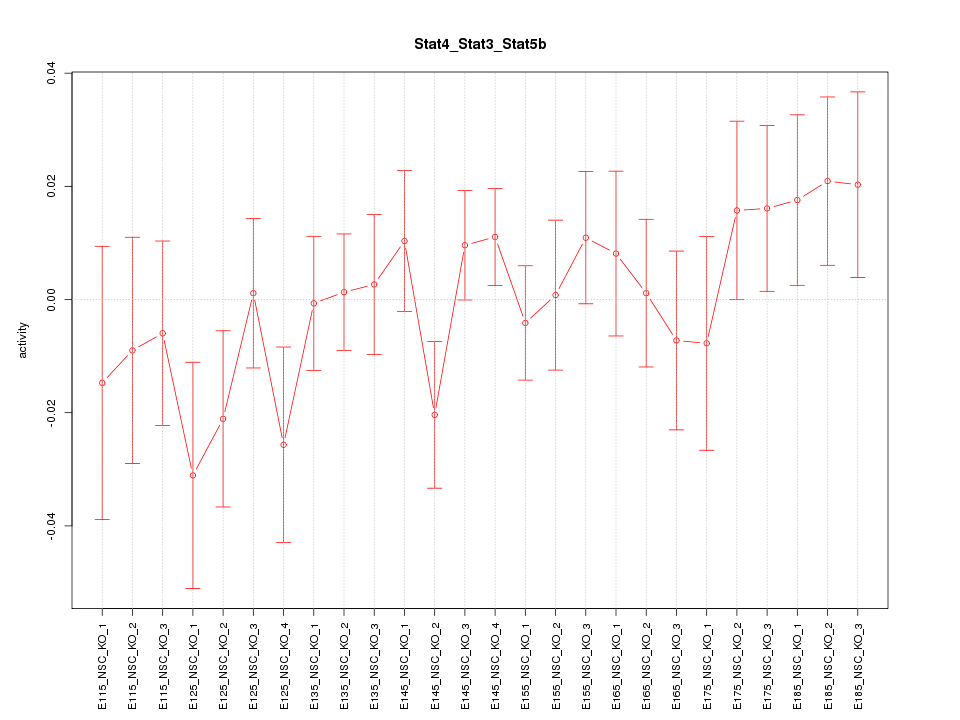
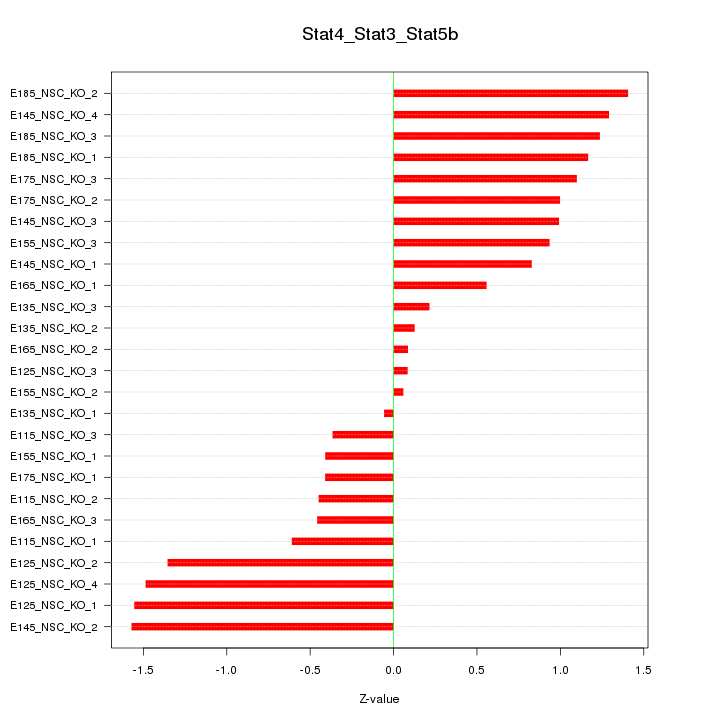
Motif ID: Stat4_Stat3_Stat5b
Z-value: 0.917

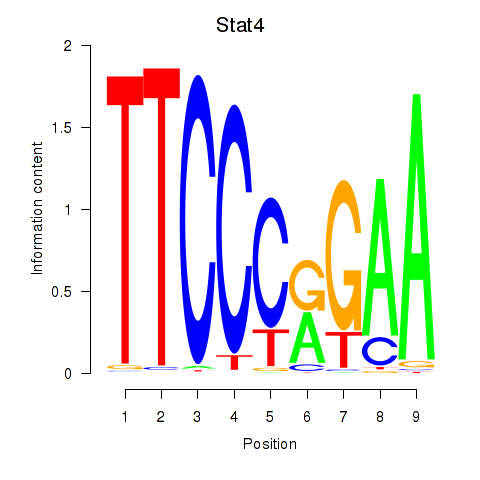
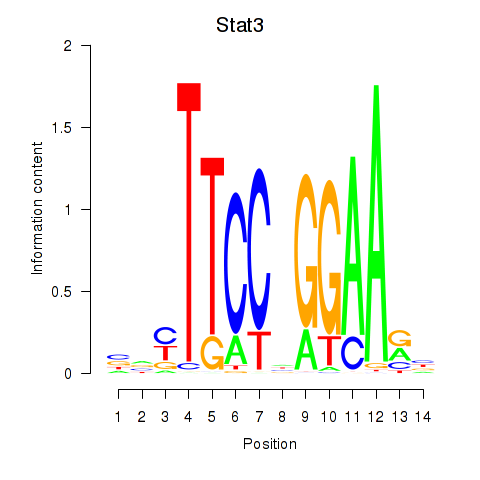
Transcription factors associated with Stat4_Stat3_Stat5b:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Stat3 | ENSMUSG00000004040.10 | Stat3 |
| Stat4 | ENSMUSG00000062939.5 | Stat4 |
| Stat5b | ENSMUSG00000020919.5 | Stat5b |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat4 | mm10_v2_chr1_+_51987139_51987151 | -0.49 | 1.1e-02 | Click! |
| Stat5b | mm10_v2_chr11_-_100822525_100822576 | -0.38 | 5.8e-02 | Click! |
| Stat3 | mm10_v2_chr11_-_100939357_100939456 | 0.31 | 1.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 1.7 | 5.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.1 | 3.4 | GO:0043379 | memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.5 | 3.2 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.5 | 1.6 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 | 1.8 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.4 | 0.4 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.4 | 1.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 1.7 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 1.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 1.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 0.8 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.2 | 6.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 5.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 1.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 0.4 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.2 | 1.6 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.2 | 2.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.5 | GO:1902524 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 3.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 0.5 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.2 | 0.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.5 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.2 | 0.6 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 1.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.5 | GO:2000569 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.9 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.4 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 0.9 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.5 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.1 | 0.8 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.4 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 | 0.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.9 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.5 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) regulation of response to drug(GO:2001023) |
| 0.1 | 0.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.9 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.1 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 1.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.3 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.2 | GO:0032242 | regulation of nucleoside transport(GO:0032242) necroptotic signaling pathway(GO:0097527) |
| 0.1 | 1.8 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.3 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 1.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 2.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.2 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 | 0.2 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.7 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.1 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 | 0.2 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.8 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.3 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.3 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.7 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 1.2 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.0 | GO:0071335 | hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.4 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 1.3 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.4 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 1.6 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.5 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.5 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 | 0.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 1.7 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0045072 | enucleate erythrocyte differentiation(GO:0043353) regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.6 | GO:0043620 | regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.5 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.1 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.0 | GO:0090285 | regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.5 | 8.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 1.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.5 | 3.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.3 | 5.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 1.0 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 0.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 5.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 0.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 2.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.3 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.8 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 2.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0033263 | HOPS complex(GO:0030897) CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 2.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0097342 | death-inducing signaling complex(GO:0031264) ripoptosome(GO:0097342) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.9 | 5.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.8 | 3.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.6 | 5.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 1.6 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.4 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 1.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 1.2 | GO:0019962 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) interleukin-20 binding(GO:0042015) |
| 0.3 | 1.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.2 | 2.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 1.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 1.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 3.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.5 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.5 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0097001 | sphingolipid transporter activity(GO:0046624) ceramide binding(GO:0097001) |
| 0.1 | 1.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.3 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.4 | GO:0050062 | long-chain-fatty-acyl-CoA reductase activity(GO:0050062) fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 2.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.1 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.6 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 6.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.0 | GO:0023026 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.7 | GO:0017166 | dystroglycan binding(GO:0002162) vinculin binding(GO:0017166) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 2.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.1 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0032052 | purine nucleobase binding(GO:0002060) bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.0 | 0.3 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.6 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.0 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |