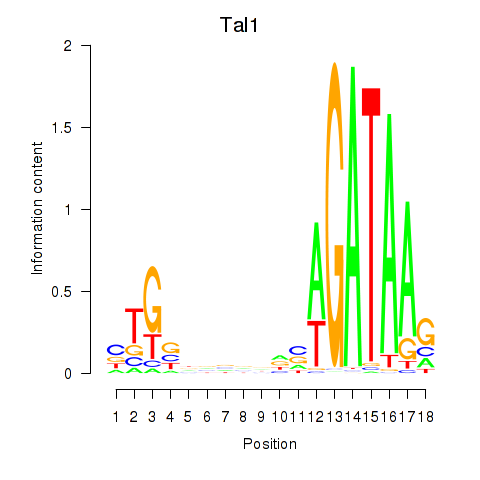
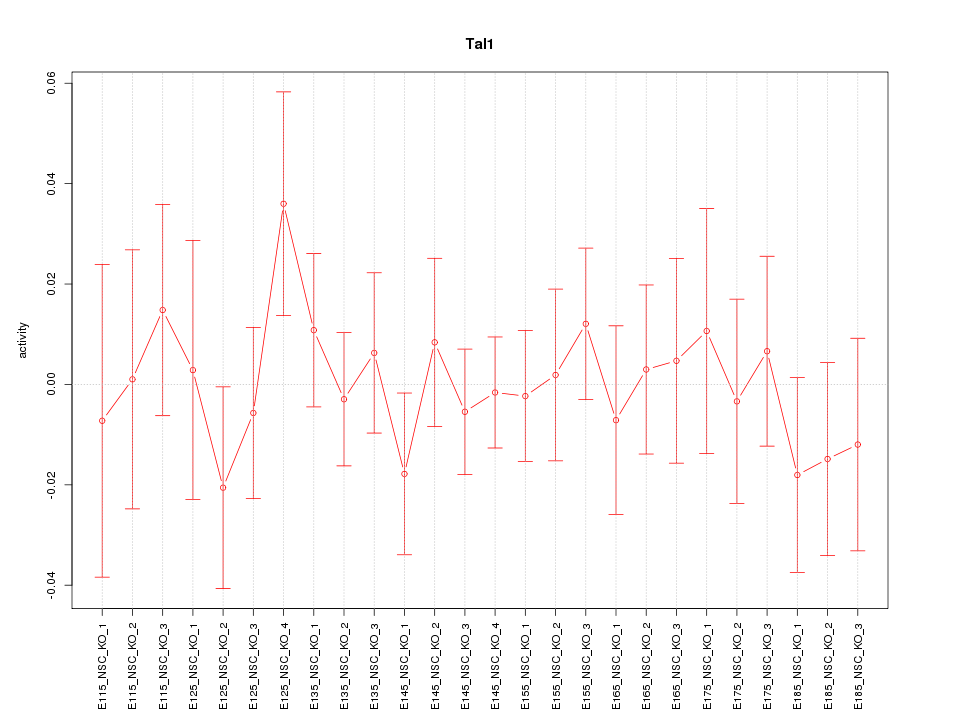
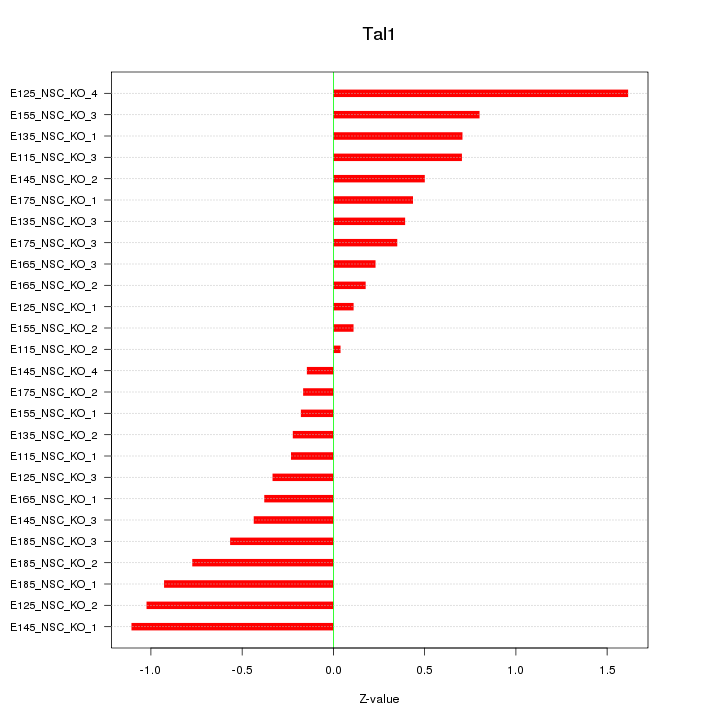
| 2.6 |
12.9 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.3 |
1.3 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.2 |
1.0 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 |
0.5 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
0.6 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.7 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.1 |
0.7 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
0.7 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.1 |
0.3 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 |
0.3 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 |
0.9 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 |
0.4 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 |
0.4 |
GO:0051096 |
telomere assembly(GO:0032202) positive regulation of helicase activity(GO:0051096) |
| 0.1 |
0.2 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 |
0.2 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.4 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.2 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.0 |
0.2 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.3 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.4 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.0 |
0.6 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 |
0.3 |
GO:0032782 |
bile acid secretion(GO:0032782) positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.2 |
GO:0000189 |
MAPK import into nucleus(GO:0000189) |
| 0.0 |
0.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.1 |
GO:0006059 |
hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.0 |
0.1 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.0 |
0.4 |
GO:0086023 |
adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 |
0.3 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.0 |
0.6 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.1 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.0 |
0.2 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.1 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.0 |
0.2 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.1 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 |
0.1 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.0 |
0.2 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.2 |
GO:0010804 |
negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 |
0.2 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 |
0.1 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.1 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 |
0.3 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.3 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
1.1 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 |
0.4 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.4 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.1 |
GO:0002098 |
tRNA wobble uridine modification(GO:0002098) I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.4 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.2 |
GO:0034377 |
plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 |
0.3 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.1 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
0.1 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |