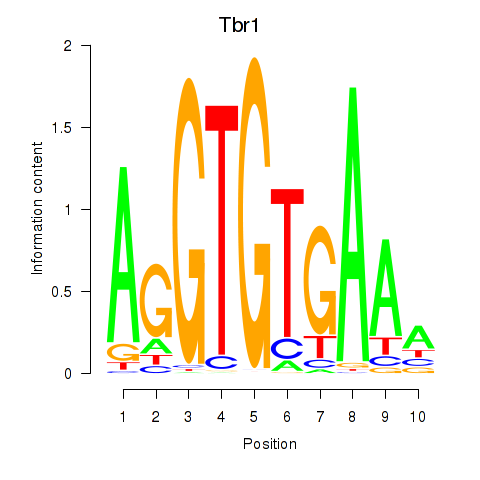
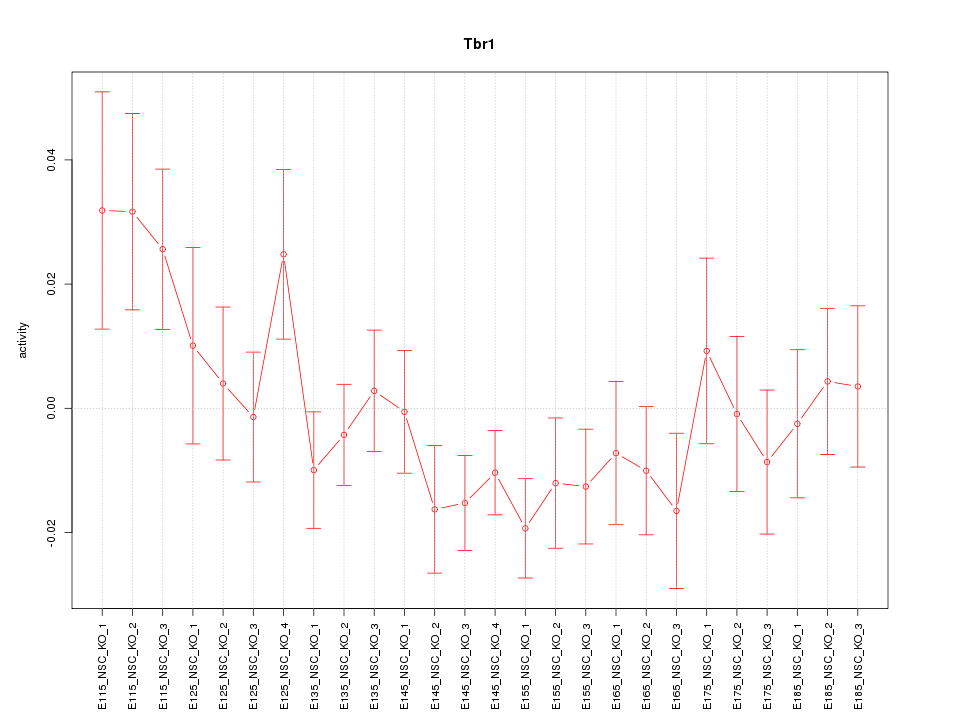
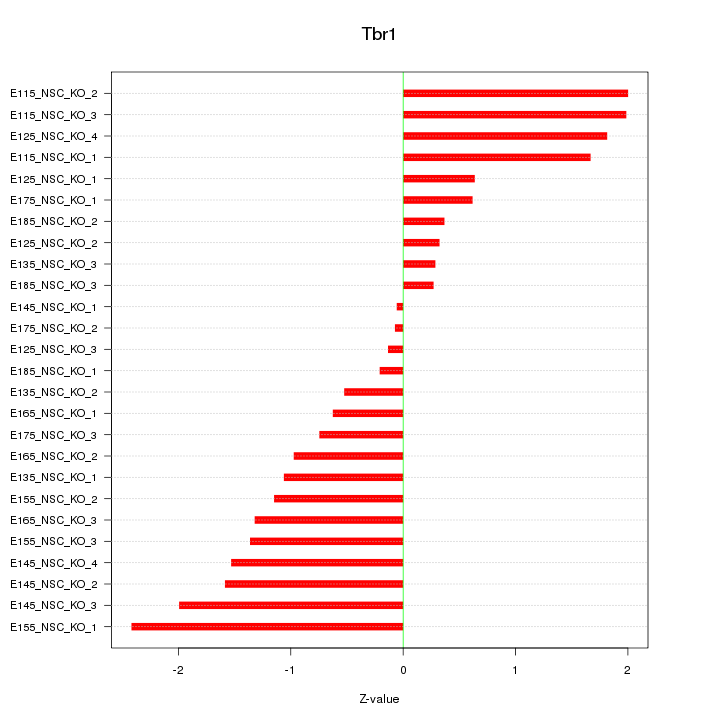
| 1.0 |
5.0 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.9 |
3.4 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.9 |
2.6 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.8 |
3.3 |
GO:0002339 |
B cell selection(GO:0002339) |
| 0.8 |
4.1 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.7 |
2.2 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.7 |
2.2 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.7 |
2.1 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.7 |
2.1 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.7 |
2.8 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.7 |
2.0 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.6 |
3.6 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.6 |
1.7 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.6 |
1.7 |
GO:0030421 |
defecation(GO:0030421) |
| 0.6 |
3.4 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.5 |
0.5 |
GO:0003134 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.5 |
1.6 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.5 |
3.8 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.5 |
1.4 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.4 |
1.3 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 |
1.7 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.4 |
1.7 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.4 |
1.6 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.4 |
1.5 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.4 |
1.1 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.4 |
1.1 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 |
1.4 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.3 |
2.1 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 |
1.0 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.3 |
1.0 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.3 |
0.3 |
GO:0035934 |
corticosterone secretion(GO:0035934) |
| 0.3 |
2.6 |
GO:0051255 |
spindle midzone assembly(GO:0051255) |
| 0.3 |
1.0 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 |
1.6 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 |
1.9 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 |
0.9 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.3 |
1.3 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.3 |
0.9 |
GO:0070366 |
regulation of hepatocyte differentiation(GO:0070366) |
| 0.3 |
3.9 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 |
1.5 |
GO:0071038 |
nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.3 |
0.9 |
GO:0090526 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 |
1.4 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.3 |
2.2 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.3 |
0.8 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 |
0.8 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.3 |
1.1 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.3 |
0.8 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.3 |
1.0 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.3 |
1.8 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 |
1.0 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 |
0.7 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.2 |
1.7 |
GO:0051461 |
positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 |
0.9 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.2 |
0.7 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.2 |
1.6 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.2 |
0.9 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.2 |
1.3 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.2 |
1.3 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
0.2 |
GO:0032811 |
negative regulation of epinephrine secretion(GO:0032811) |
| 0.2 |
0.6 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 |
0.6 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.2 |
1.5 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 |
0.6 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.2 |
1.3 |
GO:2000781 |
DNA dealkylation involved in DNA repair(GO:0006307) positive regulation of double-strand break repair(GO:2000781) |
| 0.2 |
0.6 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.2 |
1.1 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.2 |
0.6 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 |
0.5 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 |
0.7 |
GO:0070814 |
hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.2 |
1.4 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.2 |
0.7 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 |
0.9 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.2 |
0.2 |
GO:0032610 |
interleukin-1 alpha production(GO:0032610) |
| 0.2 |
0.7 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.2 |
0.7 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.2 |
0.7 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.2 |
0.5 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 |
0.5 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 |
0.5 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.2 |
3.6 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.2 |
1.8 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 |
0.3 |
GO:0002834 |
immune response to tumor cell(GO:0002418) regulation of response to tumor cell(GO:0002834) regulation of immune response to tumor cell(GO:0002837) |
| 0.2 |
0.5 |
GO:2000561 |
CD4-positive, alpha-beta T cell proliferation(GO:0035739) negative regulation of alpha-beta T cell proliferation(GO:0046642) negative regulation of interferon-gamma secretion(GO:1902714) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 |
0.5 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.2 |
0.6 |
GO:0060161 |
positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 |
1.3 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
1.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.6 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 |
0.4 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 |
1.4 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.8 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.4 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.1 |
0.4 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 |
0.4 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 |
0.3 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.1 |
1.1 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.1 |
1.3 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
3.0 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
0.9 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 |
0.6 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.4 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.1 |
0.6 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.1 |
1.4 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.1 |
0.5 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 |
0.2 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.1 |
0.4 |
GO:0050855 |
regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 |
0.8 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 |
0.3 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.7 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 |
0.8 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
0.3 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 |
0.2 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.8 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
0.2 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 |
0.3 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 |
1.0 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.7 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.1 |
3.1 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
0.3 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
1.0 |
GO:0002155 |
regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 |
0.7 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.3 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 |
1.9 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
1.2 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.1 |
0.5 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.1 |
0.6 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.1 |
0.3 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.1 |
0.7 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.8 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.4 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
1.5 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 |
0.6 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 |
0.3 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
1.3 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.1 |
0.8 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
1.2 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
1.7 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.1 |
0.3 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 |
0.9 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 |
0.3 |
GO:0015793 |
renal water transport(GO:0003097) glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.1 |
0.3 |
GO:1904338 |
positive regulation of Schwann cell migration(GO:1900149) regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 |
0.7 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.1 |
0.5 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 |
0.7 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.4 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
0.6 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 |
0.2 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.6 |
GO:0032463 |
regulation of protein homooligomerization(GO:0032462) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.7 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 |
0.5 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.9 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.1 |
0.2 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 |
0.8 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.2 |
GO:0046391 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 |
0.8 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 |
0.2 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.7 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.2 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 |
0.2 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 |
0.9 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 |
0.1 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) response to peptidoglycan(GO:0032494) |
| 0.1 |
0.8 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.2 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
0.6 |
GO:0042534 |
tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.1 |
0.6 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.3 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.1 |
0.4 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.3 |
GO:0090024 |
negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 |
0.5 |
GO:0070633 |
transepithelial transport(GO:0070633) |
| 0.1 |
0.4 |
GO:0098734 |
protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.1 |
0.2 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.6 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.1 |
0.3 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.3 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.1 |
0.8 |
GO:0021670 |
lateral ventricle development(GO:0021670) |
| 0.1 |
0.2 |
GO:0045835 |
negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 |
1.0 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
0.4 |
GO:1902475 |
ornithine transport(GO:0015822) L-alpha-amino acid transmembrane transport(GO:1902475) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 |
0.8 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 |
0.3 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.4 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.1 |
0.3 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 |
0.2 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.1 |
0.5 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 |
0.2 |
GO:2000569 |
T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 |
0.8 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.1 |
1.7 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.2 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.4 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.9 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
2.5 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 |
1.6 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.1 |
0.4 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 |
0.2 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.1 |
0.9 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.1 |
0.2 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
0.3 |
GO:1901673 |
regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 |
1.9 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.1 |
0.4 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.1 |
0.3 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.6 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.1 |
1.2 |
GO:0040036 |
regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.1 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.3 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.0 |
0.3 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.8 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.3 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
0.8 |
GO:0048643 |
positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 |
1.6 |
GO:0031050 |
dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 |
0.3 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
1.1 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.6 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
0.3 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.0 |
0.5 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
1.3 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.5 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.7 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
2.3 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.2 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.1 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.0 |
0.2 |
GO:0032753 |
positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 |
2.3 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 |
0.4 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.0 |
0.8 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
1.1 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.3 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.3 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 |
0.4 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.4 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.3 |
GO:0006532 |
aspartate biosynthetic process(GO:0006532) |
| 0.0 |
0.2 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.0 |
0.3 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
2.6 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.6 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.2 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 |
0.4 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.7 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.1 |
GO:0038108 |
negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 |
0.3 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.7 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
1.9 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.5 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 |
1.3 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 |
0.9 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.1 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.0 |
0.3 |
GO:0046500 |
S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 |
0.4 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.8 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
1.1 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.2 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.5 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.2 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.4 |
GO:0035067 |
negative regulation of histone acetylation(GO:0035067) |
| 0.0 |
0.2 |
GO:2000811 |
regulation of anoikis(GO:2000209) negative regulation of anoikis(GO:2000811) |
| 0.0 |
0.6 |
GO:0002183 |
cytoplasmic translational initiation(GO:0002183) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.3 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.1 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.0 |
0.4 |
GO:0046514 |
ceramide catabolic process(GO:0046514) |
| 0.0 |
0.4 |
GO:0006851 |
mitochondrial calcium ion transport(GO:0006851) |
| 0.0 |
0.5 |
GO:0070232 |
regulation of T cell apoptotic process(GO:0070232) |
| 0.0 |
0.5 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.2 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.0 |
0.3 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.2 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
1.2 |
GO:1902017 |
regulation of cilium assembly(GO:1902017) |
| 0.0 |
0.4 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
1.3 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 |
0.4 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.5 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.1 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.0 |
1.4 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.2 |
GO:0045740 |
positive regulation of DNA replication(GO:0045740) |
| 0.0 |
0.3 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.5 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.1 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.0 |
0.2 |
GO:0051683 |
establishment of Golgi localization(GO:0051683) |
| 0.0 |
0.3 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.7 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.7 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 |
0.1 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.1 |
GO:1903069 |
regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 |
0.2 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 |
0.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
1.5 |
GO:0001824 |
blastocyst development(GO:0001824) |
| 0.0 |
0.4 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.4 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
0.1 |
GO:0071028 |
nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.4 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.5 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
1.3 |
GO:0008584 |
male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 |
0.0 |
GO:0002385 |
organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0009136 |
ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
1.8 |
GO:0050658 |
nucleic acid transport(GO:0050657) RNA transport(GO:0050658) |
| 0.0 |
0.2 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
1.0 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.1 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.9 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.2 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.5 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.1 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.0 |
0.1 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.1 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.2 |
GO:0019731 |
antibacterial humoral response(GO:0019731) |
| 0.0 |
0.1 |
GO:0048311 |
mitochondrion distribution(GO:0048311) |
| 0.0 |
0.2 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.0 |
0.1 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.0 |
0.3 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.3 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 |
0.4 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.3 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.0 |
GO:1903894 |
regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 |
0.2 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.0 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.0 |
GO:0042255 |
ribosome assembly(GO:0042255) |
| 0.0 |
0.1 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.0 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.1 |
GO:0000460 |
maturation of 5.8S rRNA(GO:0000460) |