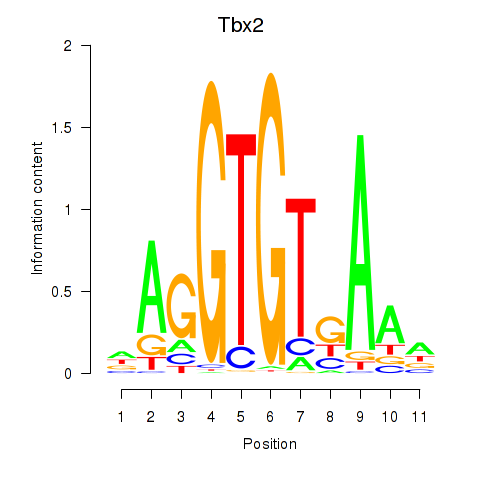
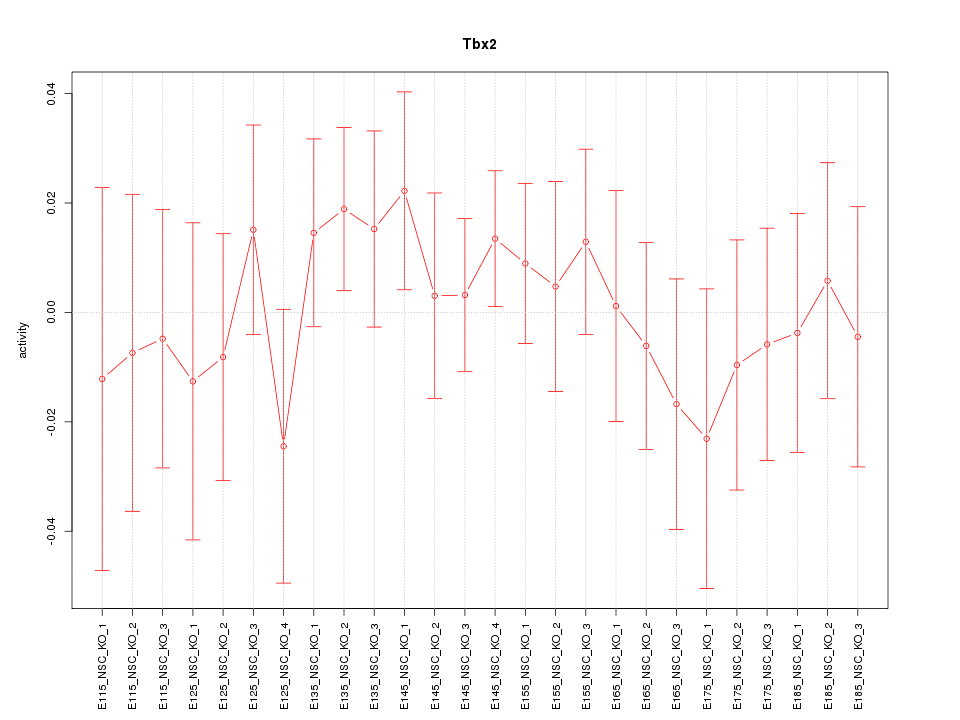
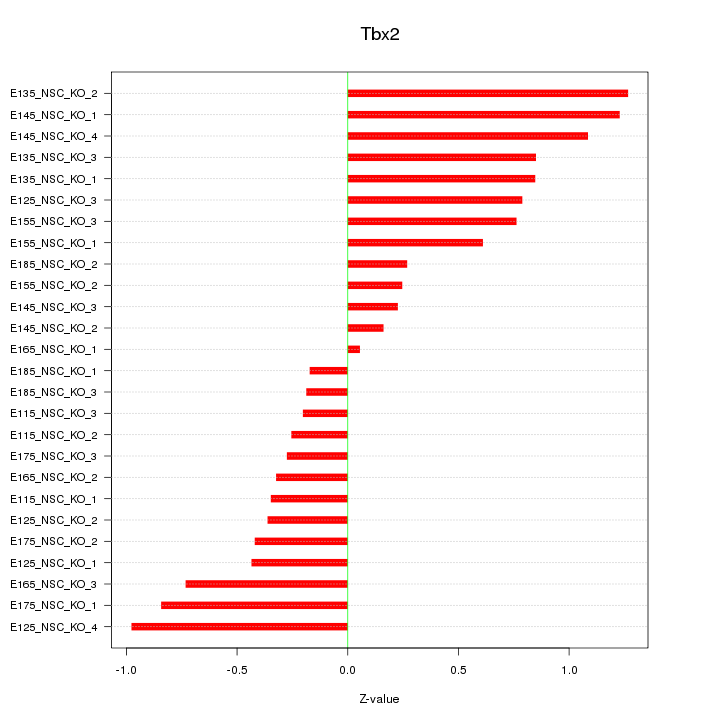
| 0.3 |
1.7 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
0.7 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 |
1.0 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 |
0.8 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.2 |
0.7 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 |
0.7 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 |
0.5 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.2 |
0.5 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.7 |
GO:0060467 |
negative regulation of fertilization(GO:0060467) |
| 0.1 |
0.7 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.1 |
0.4 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 |
0.6 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.1 |
0.3 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
1.2 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
0.5 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
2.7 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 |
0.3 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 |
0.3 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.1 |
0.2 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 |
0.5 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.4 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 |
0.2 |
GO:0033364 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.1 |
0.2 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 |
0.9 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
1.5 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 |
0.3 |
GO:0048859 |
negative regulation of epidermal cell differentiation(GO:0045605) formation of anatomical boundary(GO:0048859) |
| 0.0 |
0.1 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 |
0.2 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
1.2 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.1 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 |
0.8 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.1 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 |
0.1 |
GO:0034476 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.6 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.8 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.5 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.5 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.1 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.1 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.2 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.3 |
GO:0010107 |
potassium ion import(GO:0010107) |