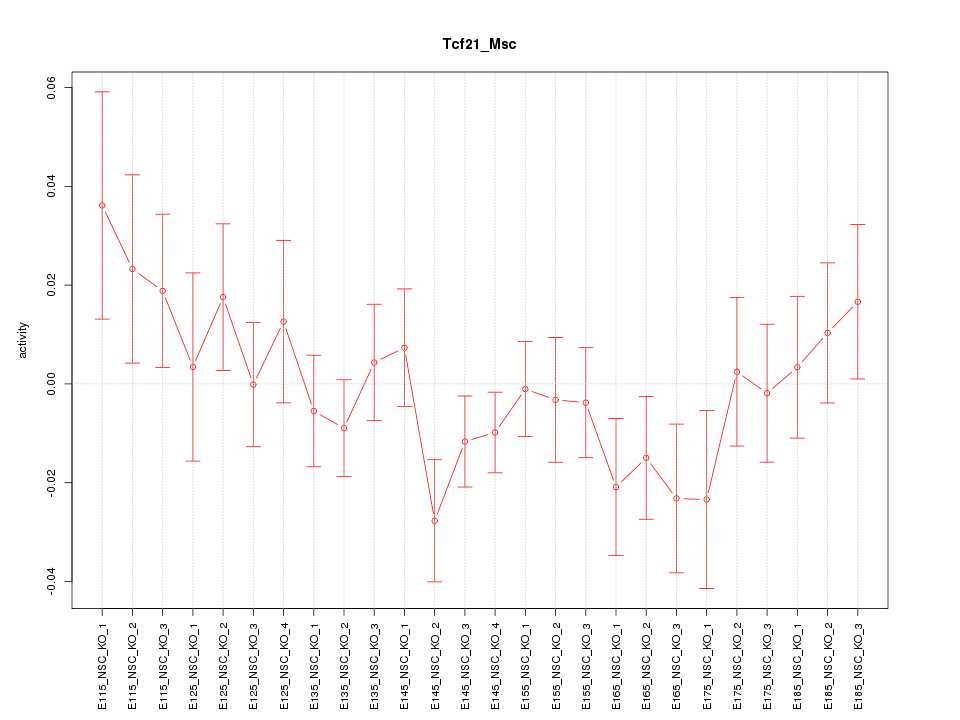
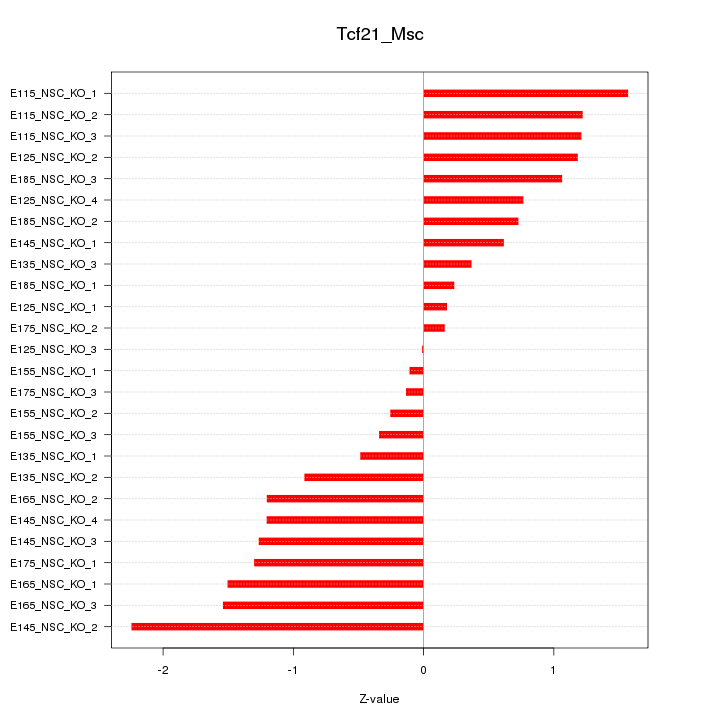
Motif ID: Tcf21_Msc
Z-value: 1.016

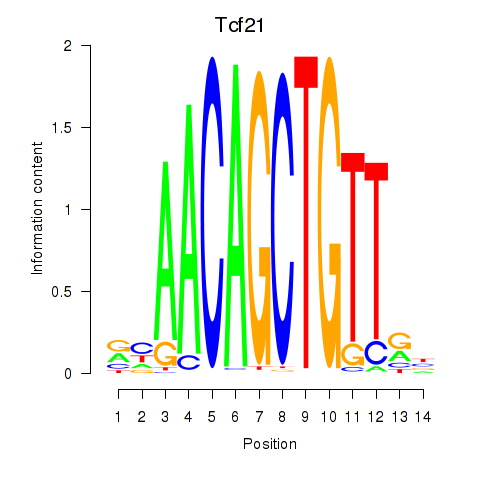
Transcription factors associated with Tcf21_Msc:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Msc | ENSMUSG00000025930.5 | Msc |
| Tcf21 | ENSMUSG00000045680.7 | Tcf21 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.0 | 3.0 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.9 | 2.6 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.6 | 2.4 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.6 | 3.6 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.6 | 1.8 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.6 | 1.8 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.6 | 1.8 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.5 | 1.6 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.4 | 1.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.4 | 2.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.4 | 1.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.4 | 1.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 2.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 | 1.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.3 | 1.0 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.3 | 1.7 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.3 | 1.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 1.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.3 | 1.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.3 | 1.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 1.3 | GO:0071205 | clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.3 | 0.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 0.3 | 2.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.0 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.2 | 0.7 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 2.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.7 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 1.6 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.2 | 2.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 0.6 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.2 | 0.9 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.2 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 1.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 0.8 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.2 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 2.3 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.2 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.7 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 0.5 | GO:1903061 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.2 | 0.5 | GO:0035799 | ureter maturation(GO:0035799) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) cell-cell signaling involved in metanephros development(GO:0072204) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.2 | 1.7 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.2 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.2 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.2 | 1.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 2.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 1.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.6 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.4 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 1.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.9 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.6 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 | 2.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.2 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 2.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.3 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.6 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 1.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 3.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.6 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.8 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 0.3 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.7 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 | 0.2 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 1.0 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 2.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.8 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 0.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.0 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.8 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 0.2 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.7 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 1.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.6 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 1.2 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.1 | 1.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.2 | GO:0006547 | histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.1 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.9 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.1 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.5 | GO:0097531 | mast cell migration(GO:0097531) |
| 0.1 | 1.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 1.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 1.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.7 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0034115 | negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.5 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 1.0 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.2 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.4 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.4 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.9 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 | 1.3 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.6 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.6 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 1.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.4 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.5 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.3 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.0 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.3 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 1.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 1.8 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.0 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 1.3 | GO:0051297 | centrosome organization(GO:0051297) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.4 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0071476 | hypotonic response(GO:0006971) cellular hypotonic response(GO:0071476) |
| 0.0 | 0.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.5 | 1.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 1.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 1.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 0.5 | GO:0030894 | replisome(GO:0030894) |
| 0.2 | 1.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.9 | GO:0001651 | dense fibrillar component(GO:0001651) box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 1.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 1.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 0.9 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.8 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 3.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.7 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.5 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 2.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 3.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 2.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 4.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 1.8 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.5 | 2.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.5 | 1.5 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 1.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.3 | 0.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 1.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 2.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.3 | 1.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 0.8 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 2.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 6.4 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 1.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 2.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 2.0 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.2 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.4 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.6 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.9 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 3.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.6 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.5 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.1 | 0.2 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.4 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 2.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.3 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.1 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.1 | 0.6 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 2.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.8 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.7 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 1.6 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.5 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.6 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 4.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 1.0 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.0 | 1.1 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 1.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.0 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.5 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-palmitoyltransferase activity(GO:0016416) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) protein-cysteine S-acyltransferase activity(GO:0019707) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) Ras palmitoyltransferase activity(GO:0043849) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 2.1 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.5 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 7.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.8 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |