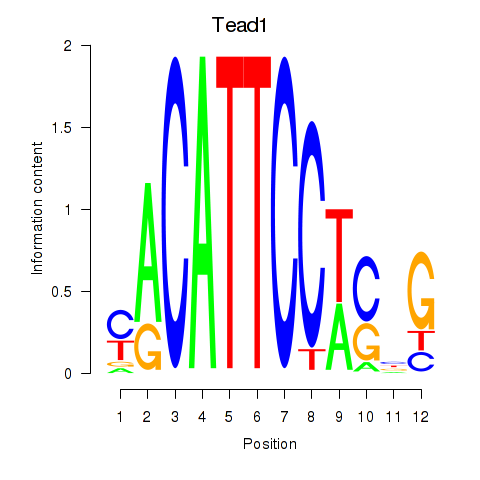
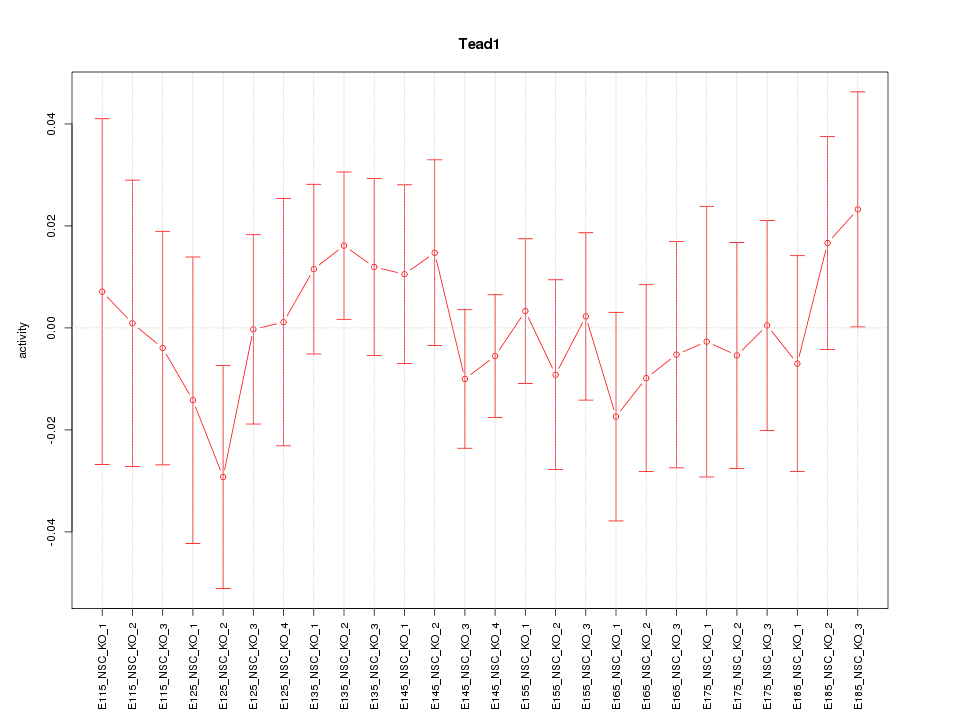
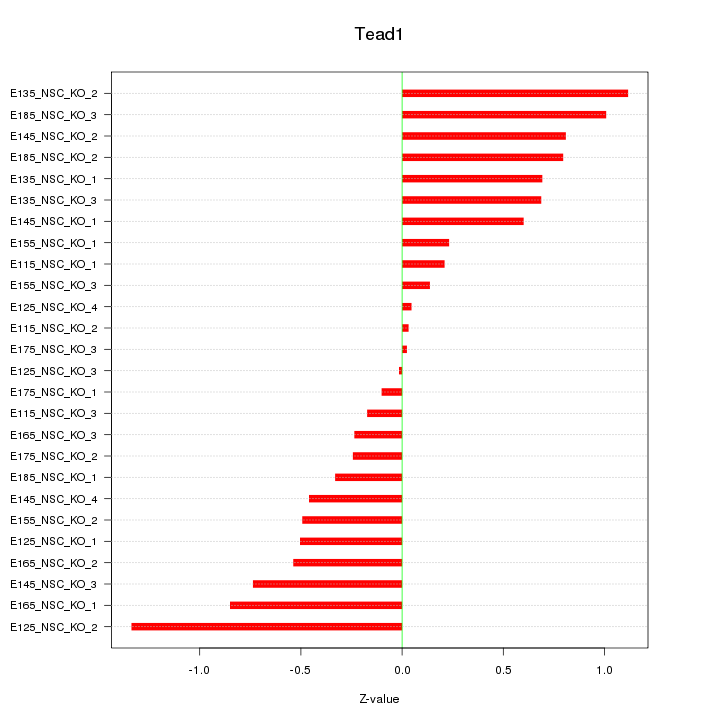
| 0.4 |
1.8 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.3 |
0.8 |
GO:1904395 |
regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 |
1.0 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.2 |
1.3 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 |
0.8 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.2 |
1.4 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 |
0.7 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 |
0.6 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.1 |
0.4 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
3.1 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.1 |
0.6 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.6 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.3 |
GO:0061324 |
mammary placode formation(GO:0060596) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.1 |
0.6 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 |
0.6 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.3 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
0.4 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.8 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 |
0.4 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 |
0.5 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.7 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 |
0.3 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.1 |
0.5 |
GO:0010759 |
positive regulation of platelet activation(GO:0010572) positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 |
0.3 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 |
1.4 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.5 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.1 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
0.2 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.9 |
GO:2000249 |
regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 |
0.0 |
GO:0070103 |
regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.0 |
0.8 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.2 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.1 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 |
1.0 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 |
0.1 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.8 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.3 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.3 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |