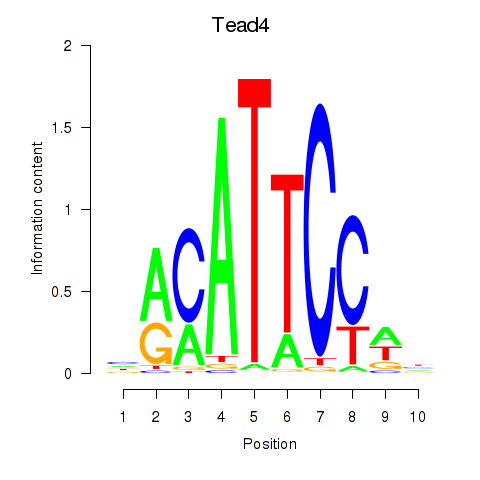
| 6.7 |
27.0 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 1.8 |
5.4 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 1.7 |
5.2 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 1.6 |
6.6 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.6 |
4.8 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 1.5 |
7.5 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.5 |
7.3 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 1.4 |
2.9 |
GO:1904395 |
positive regulation of postsynaptic membrane organization(GO:1901628) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.4 |
11.4 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.3 |
6.3 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) regulation of cortisol biosynthetic process(GO:2000064) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 1.2 |
6.0 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 1.2 |
3.5 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 1.1 |
4.4 |
GO:0046533 |
negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 1.0 |
7.3 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 1.0 |
6.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 1.0 |
5.0 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.9 |
2.8 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.9 |
4.5 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.8 |
2.4 |
GO:0061324 |
cardiac right atrium morphogenesis(GO:0003213) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.8 |
5.4 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.8 |
2.3 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.7 |
3.7 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.7 |
1.4 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.7 |
2.7 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.7 |
2.0 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.6 |
1.9 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.6 |
4.7 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.6 |
4.6 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.6 |
6.1 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.5 |
1.1 |
GO:0021943 |
formation of radial glial scaffolds(GO:0021943) |
| 0.5 |
1.1 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) |
| 0.5 |
7.4 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.5 |
1.0 |
GO:0045608 |
inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.5 |
3.6 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.4 |
0.4 |
GO:0048850 |
hypophysis morphogenesis(GO:0048850) |
| 0.4 |
1.3 |
GO:0060558 |
regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.4 |
1.3 |
GO:0014028 |
notochord formation(GO:0014028) negative regulation of lymphangiogenesis(GO:1901491) |
| 0.4 |
2.1 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.4 |
4.8 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.4 |
3.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 |
3.5 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.4 |
2.7 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 |
4.2 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.4 |
4.0 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 |
1.4 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.3 |
1.4 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.3 |
1.0 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) astrocyte activation(GO:0048143) |
| 0.3 |
2.1 |
GO:0003062 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.3 |
2.0 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.3 |
4.7 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.3 |
1.3 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.3 |
1.7 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.3 |
1.0 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.3 |
1.6 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.3 |
2.3 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.3 |
1.0 |
GO:1900158 |
regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 |
1.6 |
GO:1903598 |
negative regulation of pinocytosis(GO:0048550) angiotensin-activated signaling pathway involved in heart process(GO:0086098) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) |
| 0.3 |
3.4 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 |
6.1 |
GO:0001946 |
lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.3 |
3.0 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.3 |
0.9 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.3 |
1.7 |
GO:0035743 |
CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.3 |
2.5 |
GO:2000645 |
negative regulation of receptor catabolic process(GO:2000645) |
| 0.3 |
2.8 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 |
2.8 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.3 |
1.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 |
1.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 |
1.8 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 |
1.1 |
GO:0099624 |
regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.3 |
1.8 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.3 |
2.3 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.2 |
0.7 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 |
0.7 |
GO:0030202 |
heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.2 |
1.7 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 |
11.9 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.2 |
1.2 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 |
1.6 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 |
1.4 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.2 |
1.1 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 |
1.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 |
0.6 |
GO:2001045 |
negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 |
2.4 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 |
3.7 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.2 |
2.7 |
GO:1904293 |
negative regulation of ERAD pathway(GO:1904293) |
| 0.2 |
1.4 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 |
3.4 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.2 |
1.4 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 |
1.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.2 |
2.2 |
GO:0002467 |
germinal center formation(GO:0002467) |
| 0.2 |
0.8 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 |
0.6 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.2 |
0.6 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.2 |
2.0 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.2 |
2.4 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 |
1.2 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.1 |
1.2 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.7 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
1.3 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
5.1 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
1.3 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 |
0.3 |
GO:0060686 |
regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 |
1.6 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.1 |
0.3 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) |
| 0.1 |
2.9 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
2.4 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
1.6 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 |
1.8 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.1 |
2.8 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.1 |
0.3 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
1.0 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.1 |
0.7 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 |
0.1 |
GO:2000832 |
negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
0.4 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.3 |
GO:0090110 |
cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
0.9 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 |
0.4 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
2.4 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 |
0.4 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 |
4.5 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.1 |
3.5 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 |
3.4 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 |
1.6 |
GO:2000249 |
regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 |
1.6 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 |
4.1 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 |
0.8 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 |
1.0 |
GO:0035767 |
endothelial cell chemotaxis(GO:0035767) |
| 0.1 |
0.8 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
0.9 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
1.1 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 |
1.0 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.1 |
0.5 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
1.4 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
1.0 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.3 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) regulation of histone deacetylase activity(GO:1901725) |
| 0.0 |
0.7 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
2.9 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.3 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.3 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.3 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.0 |
4.5 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.3 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
1.7 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.5 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
5.5 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.2 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.0 |
2.1 |
GO:0042307 |
positive regulation of protein import into nucleus(GO:0042307) |
| 0.0 |
0.2 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.8 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
1.6 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.6 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 |
1.4 |
GO:0017145 |
stem cell division(GO:0017145) |
| 0.0 |
0.5 |
GO:1903846 |
positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 |
2.6 |
GO:0070374 |
positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 |
1.0 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.8 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.1 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 |
0.7 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 |
0.2 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 |
0.3 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
1.4 |
GO:0050728 |
negative regulation of inflammatory response(GO:0050728) |
| 0.0 |
1.8 |
GO:0045727 |
positive regulation of translation(GO:0045727) |
| 0.0 |
0.6 |
GO:0031076 |
embryonic camera-type eye development(GO:0031076) |
| 0.0 |
0.6 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
1.7 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.6 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.1 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 |
0.1 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.0 |
0.3 |
GO:0006006 |
glucose metabolic process(GO:0006006) |
| 0.0 |
0.4 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
0.4 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.2 |
GO:0036093 |
male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 |
0.5 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.6 |
GO:0048747 |
muscle fiber development(GO:0048747) |
| 0.0 |
1.0 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.0 |
0.3 |
GO:0016572 |
histone phosphorylation(GO:0016572) |
| 0.0 |
0.7 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.0 |
0.2 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.1 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.7 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.5 |
GO:0032543 |
mitochondrial translation(GO:0032543) |