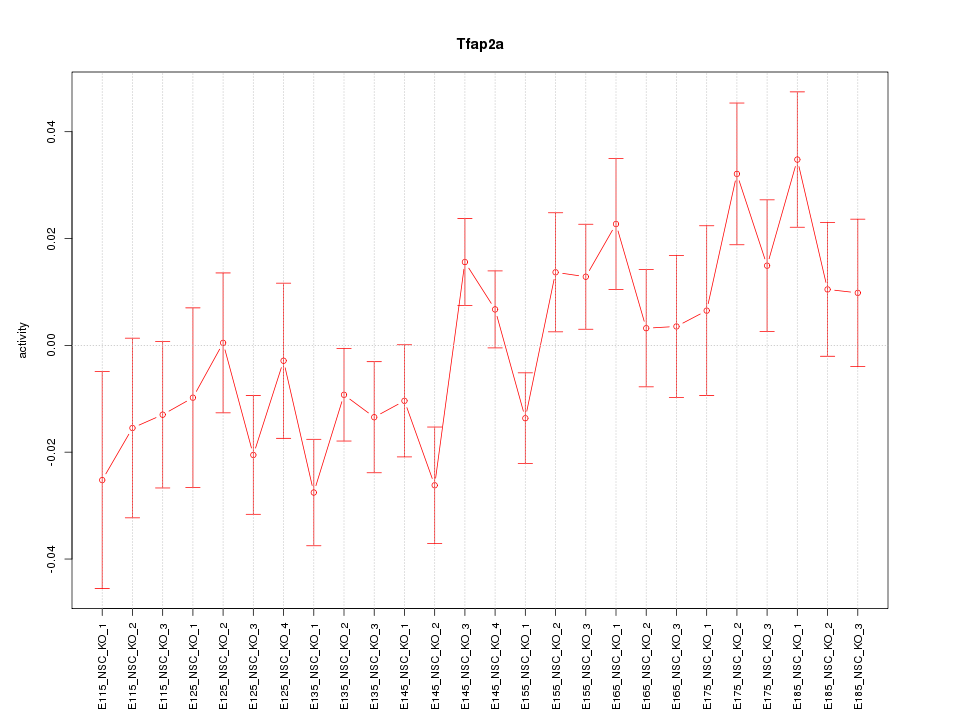
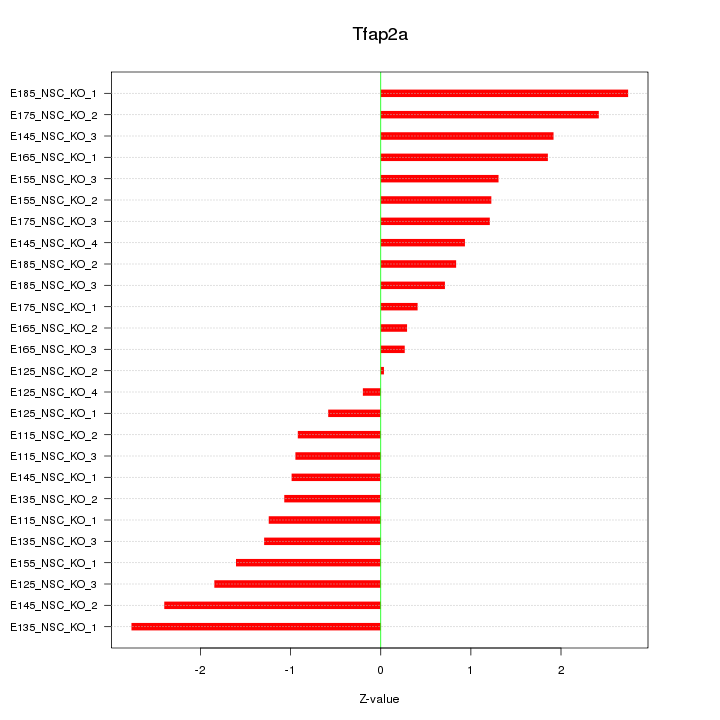
| 1.9 |
7.6 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 1.7 |
5.0 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.4 |
6.9 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 1.2 |
15.6 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.9 |
3.7 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.9 |
4.4 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.8 |
2.3 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.7 |
2.2 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.7 |
4.2 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.6 |
3.2 |
GO:0034441 |
plasma lipoprotein particle oxidation(GO:0034441) |
| 0.6 |
1.9 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.6 |
2.5 |
GO:0033024 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.6 |
1.7 |
GO:0046959 |
habituation(GO:0046959) |
| 0.5 |
1.5 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.5 |
1.9 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 |
3.8 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.4 |
2.2 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.4 |
2.2 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.4 |
1.3 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.4 |
1.2 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.4 |
1.5 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 |
1.9 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 |
1.0 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.3 |
6.8 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.3 |
0.7 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.3 |
1.9 |
GO:0000820 |
regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.3 |
1.5 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 |
0.9 |
GO:1990314 |
pyrimidine-containing compound transmembrane transport(GO:0072531) cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.3 |
1.8 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 |
0.9 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 |
0.8 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.3 |
0.8 |
GO:0010752 |
signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.3 |
1.0 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.3 |
1.8 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.3 |
1.0 |
GO:0002175 |
protein localization to paranode region of axon(GO:0002175) |
| 0.2 |
0.7 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.2 |
2.7 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 |
1.1 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.2 |
2.2 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 |
1.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 |
0.6 |
GO:0061535 |
glutamate secretion, neurotransmission(GO:0061535) |
| 0.2 |
1.3 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 |
1.9 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.2 |
2.8 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.2 |
0.6 |
GO:0009182 |
purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.2 |
0.7 |
GO:0010046 |
arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.2 |
8.4 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.2 |
1.2 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 |
0.9 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 |
0.3 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.2 |
0.7 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 |
0.8 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.2 |
0.8 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 |
0.5 |
GO:0070625 |
zymogen granule exocytosis(GO:0070625) |
| 0.1 |
0.4 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
2.0 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.1 |
2.3 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.1 |
1.0 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.4 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.5 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.1 |
2.9 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
1.3 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.8 |
GO:0002591 |
antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.1 |
0.9 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 |
0.5 |
GO:0010985 |
negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 |
0.8 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.1 |
0.8 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 |
0.6 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.1 |
0.9 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 |
2.0 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
0.4 |
GO:0033239 |
negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.1 |
0.6 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 |
3.2 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.8 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 |
0.6 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.1 |
1.9 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.1 |
0.4 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.6 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.1 |
0.4 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 |
0.6 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 |
0.7 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.1 |
1.7 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 |
1.0 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.4 |
GO:0036216 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 |
1.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.2 |
GO:1904192 |
cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 |
1.1 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.1 |
0.6 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.4 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 |
0.7 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.5 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.1 |
2.3 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.1 |
0.5 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 |
1.3 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.1 |
2.3 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.1 |
0.2 |
GO:1903351 |
response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of membrane tubulation(GO:1903525) |
| 0.1 |
0.3 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.8 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
0.3 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 |
0.3 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.1 |
1.4 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.5 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 |
1.7 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 |
0.3 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
0.3 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.1 |
0.3 |
GO:0001844 |
protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 |
0.5 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
0.3 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 |
0.5 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.1 |
0.2 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
1.0 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.7 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
1.0 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.8 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.7 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.1 |
1.5 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 |
2.1 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 |
1.2 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.1 |
1.0 |
GO:2000300 |
regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 |
0.3 |
GO:0019885 |
antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 |
0.5 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.2 |
GO:0035964 |
vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
0.5 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.0 |
1.5 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.5 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.4 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.1 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 |
0.3 |
GO:1903551 |
regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.3 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 |
0.2 |
GO:0033499 |
galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process through glucose-1-phosphate(GO:0061622) glycolytic process from galactose(GO:0061623) |
| 0.0 |
0.4 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.7 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.2 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 |
2.7 |
GO:0017157 |
regulation of exocytosis(GO:0017157) |
| 0.0 |
0.8 |
GO:0006047 |
UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 |
0.2 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
1.9 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 |
0.2 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
1.4 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.6 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.6 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 |
0.3 |
GO:0033005 |
positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.2 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.0 |
1.1 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 |
0.8 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.5 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.1 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.0 |
1.0 |
GO:0090280 |
positive regulation of calcium ion import(GO:0090280) |
| 0.0 |
0.2 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.7 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.0 |
0.4 |
GO:0061014 |
positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 |
0.2 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.2 |
GO:0032796 |
uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.0 |
0.7 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.1 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.0 |
0.7 |
GO:2000772 |
regulation of cellular senescence(GO:2000772) |
| 0.0 |
0.5 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
0.3 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.0 |
0.4 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.1 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
2.6 |
GO:0099643 |
neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 |
0.7 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.1 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.4 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.8 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
2.2 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.0 |
0.7 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
1.1 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
1.0 |
GO:0006509 |
membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 |
0.1 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 |
0.7 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 |
0.7 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 |
0.6 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
2.5 |
GO:0008203 |
cholesterol metabolic process(GO:0008203) |
| 0.0 |
0.6 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.1 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.1 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.2 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.6 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.5 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
1.1 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.6 |
GO:0009247 |
glycolipid biosynthetic process(GO:0009247) |
| 0.0 |
0.1 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.3 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.9 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
3.2 |
GO:0006874 |
cellular calcium ion homeostasis(GO:0006874) |
| 0.0 |
0.1 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.1 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.5 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.0 |
GO:0060971 |
embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 |
0.3 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.0 |
1.5 |
GO:0051924 |
regulation of calcium ion transport(GO:0051924) |
| 0.0 |
0.6 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.1 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.2 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.6 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.2 |
GO:0014002 |
astrocyte development(GO:0014002) |
| 0.0 |
0.4 |
GO:2000171 |
negative regulation of dendrite development(GO:2000171) |
| 0.0 |
0.6 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.0 |
0.2 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
1.0 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.4 |
GO:0042775 |
mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 |
0.2 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.1 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.7 |
GO:0006694 |
steroid biosynthetic process(GO:0006694) |
| 0.0 |
0.5 |
GO:0071514 |
genetic imprinting(GO:0071514) |
| 0.0 |
0.8 |
GO:0006497 |
protein lipidation(GO:0006497) |
| 0.0 |
0.6 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
0.0 |
GO:0050856 |
regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 |
0.6 |
GO:0009142 |
nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 |
0.6 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
0.7 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |