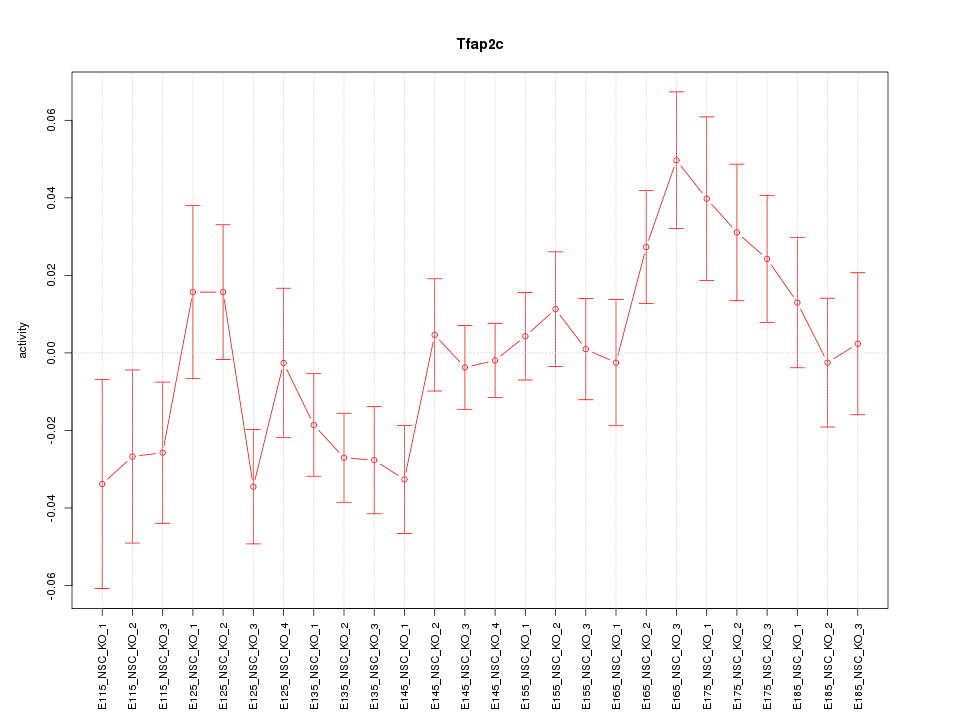
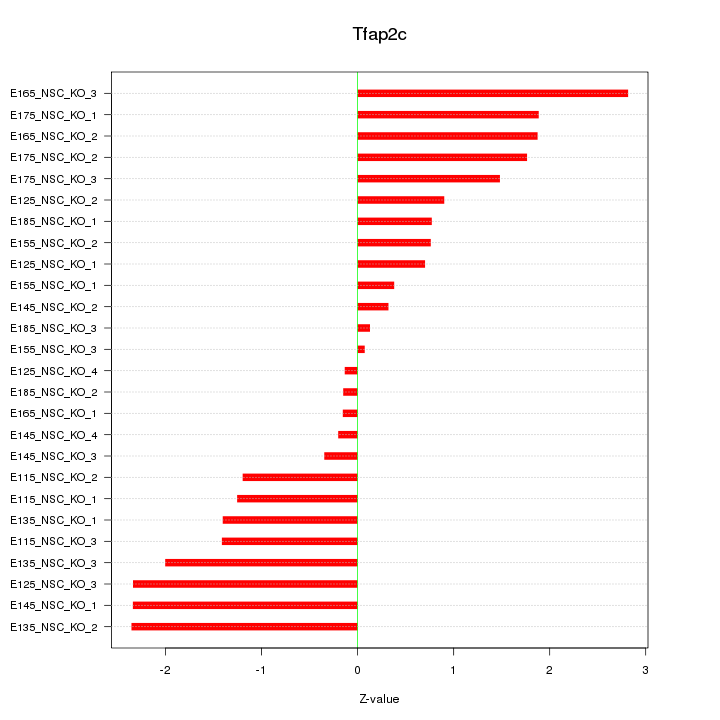
Motif ID: Tfap2c
Z-value: 1.398

Transcription factors associated with Tfap2c:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfap2c | ENSMUSG00000028640.5 | Tfap2c |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2c | mm10_v2_chr2_+_172550761_172550782 | -0.67 | 2.0e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.2 | 4.9 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 1.1 | 6.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.0 | 5.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.9 | 2.7 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) |
| 0.8 | 10.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.7 | 3.6 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.7 | 1.4 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.7 | 2.1 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.7 | 2.0 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.7 | 2.0 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.6 | 1.8 | GO:0003195 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.6 | 2.4 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.6 | 3.0 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.6 | 2.3 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 1.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.5 | 1.9 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.5 | 3.7 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.5 | 2.7 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.5 | 1.4 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.5 | 1.4 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.5 | 1.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.4 | 1.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) lateral motor column neuron migration(GO:0097477) |
| 0.4 | 1.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 1.9 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.4 | 1.5 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.4 | 1.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 3.8 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.3 | 2.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 1.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.3 | 2.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 2.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.3 | 1.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 2.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 3.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 1.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 0.8 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 1.0 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.2 | 1.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 1.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 6.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 0.5 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.2 | 0.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 1.0 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.2 | 1.1 | GO:2000348 | protein linear polyubiquitination(GO:0097039) regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.6 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.7 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.9 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.8 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 0.4 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 1.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 2.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.6 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 5.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 2.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.6 | GO:0035405 | regulation of germinal center formation(GO:0002634) histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 1.4 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.2 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.1 | 0.3 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.1 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 4.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 4.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 8.5 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 1.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.8 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 4.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 1.6 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 1.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.2 | GO:0042523 | regulation of tyrosine phosphorylation of Stat5 protein(GO:0042522) positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 2.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.9 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.4 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.9 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 2.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 2.3 | 6.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.7 | 4.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 2.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 1.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 1.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 7.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 1.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 1.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 2.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 2.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 3.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 2.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 2.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 2.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 10.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 3.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 7.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 3.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 8.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 4.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 4.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 2.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.9 | 2.7 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.9 | 2.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.7 | 9.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 3.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.6 | 2.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.6 | 3.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.6 | 2.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.6 | 2.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 2.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.4 | 2.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 1.4 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.3 | 1.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.3 | 1.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 3.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 1.0 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 0.9 | GO:0080084 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.2 | 2.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.7 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 1.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 3.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.6 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 5.8 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.2 | 4.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 2.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 1.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 2.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.6 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 1.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.6 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 1.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.5 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.1 | 6.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 1.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 7.5 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.6 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 6.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 3.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 5.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 7.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.4 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 3.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 1.0 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 4.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |