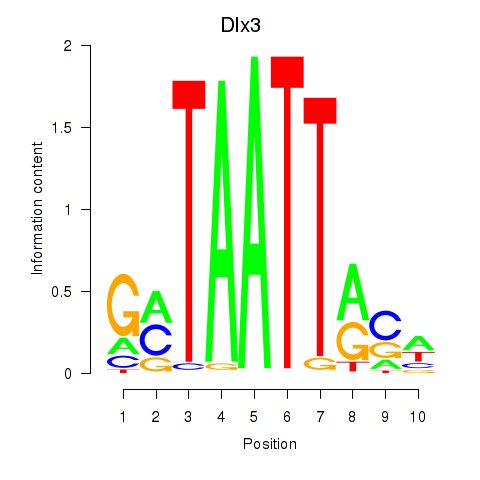
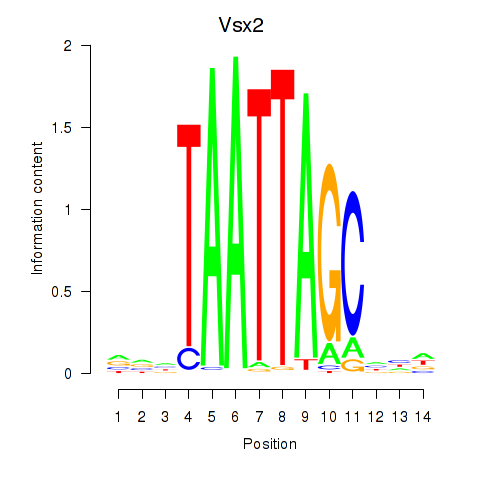
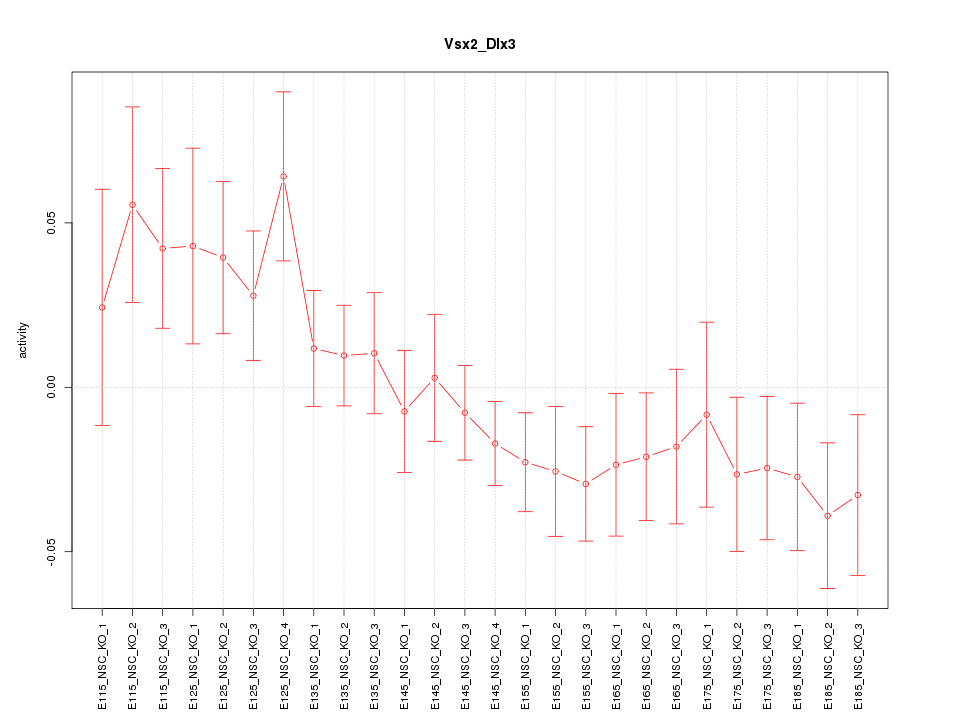
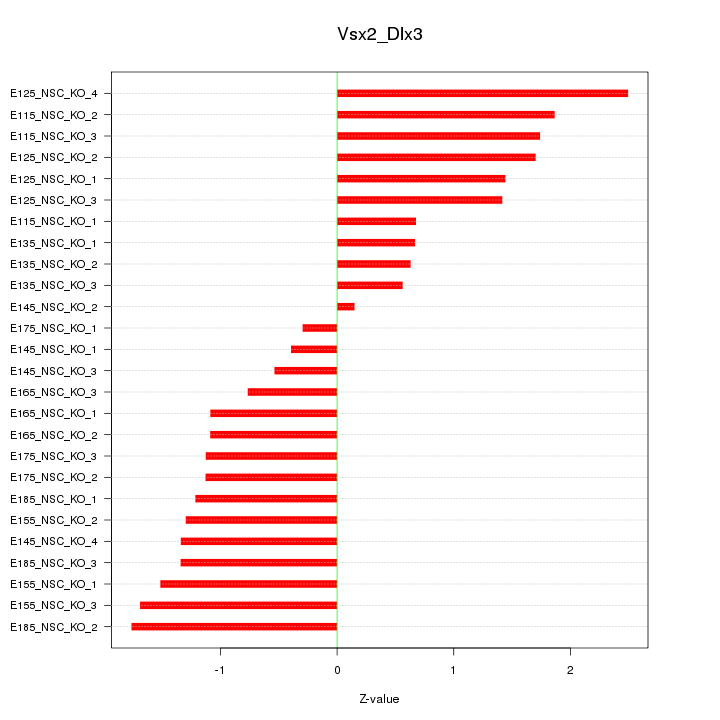
| 3.9 |
30.9 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 3.2 |
19.5 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.7 |
5.2 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 1.5 |
5.9 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.0 |
18.4 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 1.0 |
2.9 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.9 |
2.6 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.6 |
4.5 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.6 |
7.5 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.5 |
1.6 |
GO:0090526 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 |
1.3 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.3 |
1.7 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.3 |
5.0 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 |
2.2 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 |
0.9 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.3 |
5.1 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.3 |
0.8 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 |
1.2 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.2 |
0.6 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 |
0.5 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
0.6 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 |
2.5 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 |
1.9 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
1.9 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
2.0 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
5.4 |
GO:0021884 |
forebrain neuron development(GO:0021884) |
| 0.1 |
0.8 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.3 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 |
2.1 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
0.6 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.1 |
2.7 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
2.4 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.1 |
0.8 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.3 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 |
1.0 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.7 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
2.7 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.1 |
1.3 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 |
0.2 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 |
1.2 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
3.6 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
1.6 |
GO:0061028 |
establishment of endothelial barrier(GO:0061028) |
| 0.0 |
0.8 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
1.7 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.2 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.3 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.2 |
GO:0032782 |
urea transport(GO:0015840) bile acid secretion(GO:0032782) urea transmembrane transport(GO:0071918) |
| 0.0 |
0.1 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.0 |
0.2 |
GO:0090031 |
positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 |
1.2 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
1.6 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
1.5 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.4 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
1.0 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
2.2 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.2 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 |
1.0 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
1.6 |
GO:0014065 |
phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 |
0.3 |
GO:0006101 |
tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 |
2.6 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.4 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |