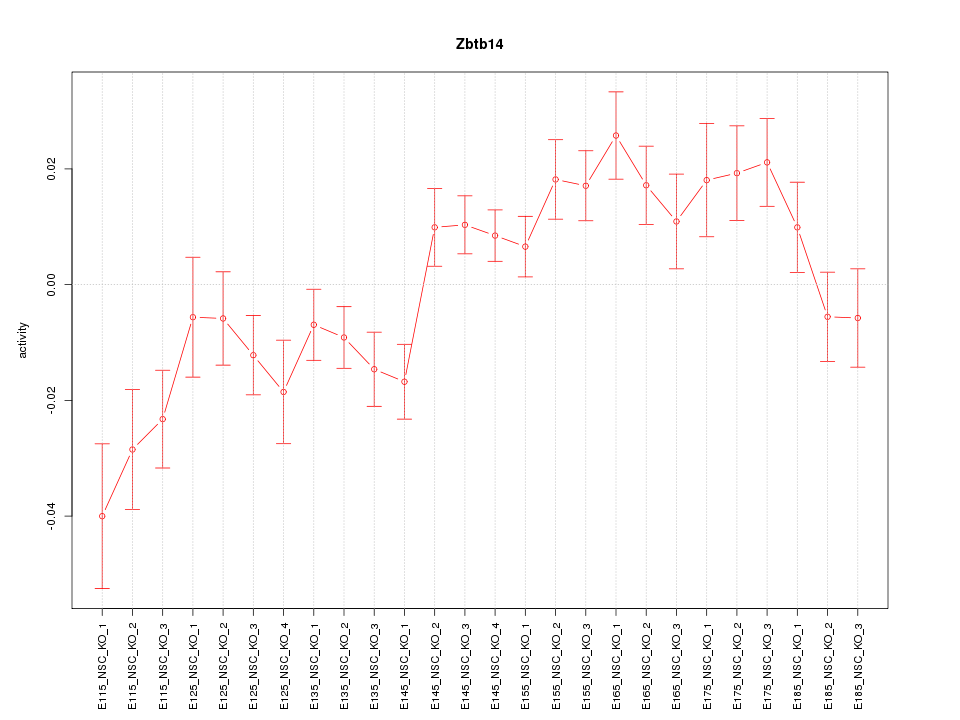
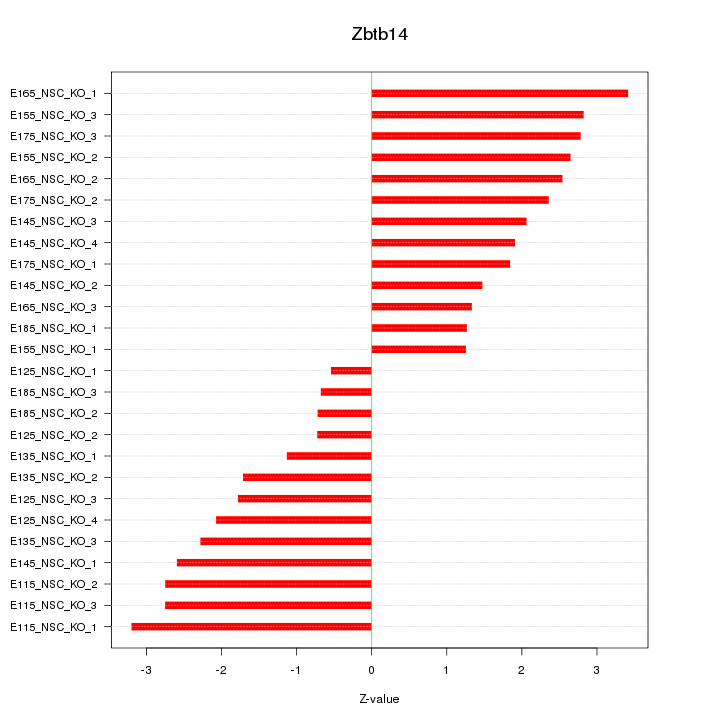
Motif ID: Zbtb14
Z-value: 2.109
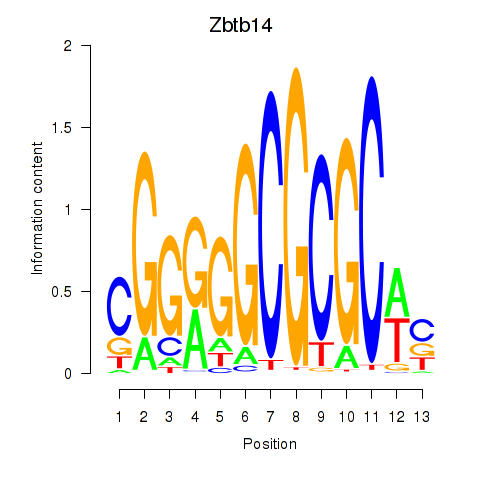
Transcription factors associated with Zbtb14:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb14 | ENSMUSG00000049672.8 | Zbtb14 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb14 | mm10_v2_chr17_+_69383319_69383394 | -0.60 | 1.3e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.6 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 3.2 | 19.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.8 | 11.3 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 2.7 | 43.5 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 2.6 | 7.7 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 2.5 | 9.8 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 2.1 | 6.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.8 | 3.6 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) |
| 1.8 | 5.3 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.7 | 5.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.7 | 5.0 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 1.6 | 4.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.4 | 4.3 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.4 | 4.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.4 | 4.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.3 | 4.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 1.3 | 5.3 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.3 | 2.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.3 | 7.8 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.2 | 3.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.2 | 5.9 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 1.2 | 3.5 | GO:0021546 | rhombomere development(GO:0021546) |
| 1.1 | 3.4 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 1.1 | 4.5 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 1.1 | 8.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.1 | 3.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 1.1 | 4.4 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 1.0 | 2.1 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 1.0 | 4.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.0 | 2.0 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 1.0 | 3.8 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.9 | 2.8 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.9 | 1.7 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.9 | 4.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.8 | 5.9 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.8 | 2.5 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.7 | 3.0 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.7 | 1.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.7 | 10.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.7 | 2.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.7 | 2.8 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.7 | 1.4 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.7 | 7.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.7 | 2.0 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.7 | 9.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 | 7.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 1.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.6 | 2.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.6 | 0.6 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.6 | 1.8 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.6 | 1.2 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.6 | 0.6 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.6 | 2.4 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.6 | 8.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.6 | 2.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.6 | 1.7 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.6 | 1.7 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.6 | 15.2 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.5 | 1.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 2.2 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.5 | 16.7 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.5 | 0.5 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.5 | 2.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.5 | 3.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 3.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.5 | 2.1 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.5 | 2.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.5 | 1.5 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.5 | 3.9 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.5 | 0.5 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.5 | 2.9 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.5 | 2.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.5 | 2.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.5 | 1.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 2.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.4 | 3.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 0.9 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.4 | 3.8 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.4 | 4.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 5.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.4 | 1.2 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) |
| 0.4 | 4.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 3.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 2.0 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.4 | 2.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 0.8 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.4 | 1.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 1.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 1.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.4 | 3.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.4 | 1.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.4 | 14.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.4 | 4.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 1.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.4 | 0.7 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.4 | 1.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.4 | 6.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 3.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 3.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 0.7 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.3 | 1.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.3 | 1.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.3 | 1.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.3 | 1.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.3 | 6.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.3 | 2.0 | GO:1902661 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.3 | 1.3 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.3 | 1.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 1.9 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.3 | 6.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 | 2.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.3 | 1.6 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.3 | 3.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 0.9 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.3 | 1.2 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.3 | 0.9 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.3 | 0.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 0.9 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.3 | 0.9 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.3 | 0.6 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.3 | 1.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.3 | 1.5 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 4.6 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.3 | 5.5 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.3 | 0.3 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.3 | 0.8 | GO:0030862 | regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.3 | 1.6 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.3 | 2.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.3 | 1.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 5.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 3.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 2.3 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.3 | 1.8 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.3 | 2.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 7.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 1.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.9 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 0.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 2.8 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 1.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 0.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 0.9 | GO:0010637 | diet induced thermogenesis(GO:0002024) negative regulation of mitochondrial fusion(GO:0010637) adaptive thermogenesis(GO:1990845) |
| 0.2 | 2.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 1.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.4 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.2 | 1.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 1.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.5 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.6 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.2 | 1.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 0.6 | GO:0060214 | stem cell fate commitment(GO:0048865) stem cell fate specification(GO:0048866) endocardium formation(GO:0060214) |
| 0.2 | 4.6 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 0.6 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.2 | 1.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 1.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 4.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.0 | GO:0044829 | negative regulation by host of viral process(GO:0044793) negative regulation by host of viral genome replication(GO:0044828) positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 4.0 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.2 | 3.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 4.0 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.2 | 0.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.2 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 2.7 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 | 0.7 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 2.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 1.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 4.3 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.2 | 0.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 0.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 2.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 3.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 0.3 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 1.5 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 2.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 0.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 1.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 0.6 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 0.8 | GO:0043314 | negative regulation of cellular extravasation(GO:0002692) negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 3.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.5 | GO:2000620 | regulation of histone H4-K16 acetylation(GO:2000618) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 5.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 2.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 0.9 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 0.2 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 1.5 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.7 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.3 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.1 | 0.9 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 1.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 3.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 1.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.6 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 4.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.5 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 2.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 1.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 4.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 1.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.4 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.3 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 0.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 1.3 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 3.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.4 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.6 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 1.8 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 6.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 7.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.7 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 1.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 1.5 | GO:0010574 | regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 1.0 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.7 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 1.1 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.1 | 0.6 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.1 | 7.6 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.6 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.7 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 2.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.5 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.1 | GO:0072610 | interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.4 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 2.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.3 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 0.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.7 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.1 | 0.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.1 | 0.3 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.6 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 | 3.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.5 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 0.5 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 1.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.8 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.3 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.3 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 2.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 3.4 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 1.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 1.5 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 3.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.8 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 5.7 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.1 | 0.4 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 1.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 1.8 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:1901071 | N-acetylglucosamine metabolic process(GO:0006044) glucosamine-containing compound metabolic process(GO:1901071) |
| 0.0 | 0.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 3.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.4 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.4 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.0 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.0 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 3.4 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 2.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0019377 | glycolipid catabolic process(GO:0019377) |
| 0.0 | 0.3 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.7 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.8 | GO:0019217 | regulation of fatty acid metabolic process(GO:0019217) |
| 0.0 | 0.2 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.1 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.8 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.1 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.0 | GO:0010273 | detoxification of copper ion(GO:0010273) detoxification of inorganic compound(GO:0061687) stress response to copper ion(GO:1990169) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.9 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.5 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.3 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0014904 | myotube cell development(GO:0014904) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 37.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 3.8 | 3.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 2.3 | 9.3 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 2.2 | 13.2 | GO:0008091 | spectrin(GO:0008091) |
| 1.7 | 6.8 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.4 | 7.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.2 | 5.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.9 | 3.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.8 | 9.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.7 | 22.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.7 | 7.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.6 | 3.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.6 | 7.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.6 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 7.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 5.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 2.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.5 | 6.2 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.5 | 1.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 4.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 1.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.4 | 1.3 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.4 | 7.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 1.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 6.2 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.4 | 2.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 10.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.4 | 1.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.4 | 22.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 5.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.3 | 2.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 3.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 0.9 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.3 | 2.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 1.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 2.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 8.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 0.9 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 0.4 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.2 | 2.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.2 | 1.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 24.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 3.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 2.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.9 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 5.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 1.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 5.4 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 8.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.3 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.1 | 4.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 14.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 4.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.6 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 5.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.5 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 3.3 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.1 | 0.8 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 4.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 6.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 5.1 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 3.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 11.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 12.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 1.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 1.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 4.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.0 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 2.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 8.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.6 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 8.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 2.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.8 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 1.9 | 15.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.9 | 9.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 1.8 | 5.3 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 1.5 | 7.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.5 | 8.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.5 | 33.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.5 | 8.8 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.3 | 6.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.1 | 3.4 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.9 | 2.8 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.9 | 2.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.9 | 3.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 21.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.9 | 7.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.8 | 8.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.8 | 4.7 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.8 | 1.5 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.7 | 4.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 2.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.7 | 4.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.7 | 2.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.7 | 4.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.6 | 9.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.6 | 12.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.6 | 5.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.6 | 1.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.6 | 3.4 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.6 | 13.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.6 | 6.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.6 | 3.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 1.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.5 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 8.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.5 | 5.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.5 | 4.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 1.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.4 | 3.8 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.4 | 2.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 5.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 2.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 1.6 | GO:0008061 | chitin binding(GO:0008061) |
| 0.4 | 1.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 2.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 8.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 1.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 3.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 2.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 2.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 1.0 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.3 | 2.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 0.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.3 | 2.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 3.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 0.9 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.3 | 0.9 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.3 | 1.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 9.3 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.3 | 2.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 1.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 1.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 1.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.3 | 3.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 3.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 3.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 9.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 3.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 10.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 4.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 0.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 3.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.9 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 5.0 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 2.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.2 | 3.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 2.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 2.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 3.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 9.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 2.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 10.7 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.2 | 1.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 5.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.2 | 6.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 1.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 2.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 2.1 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.2 | 5.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 4.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 0.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 6.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 2.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 2.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.0 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.1 | 0.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 5.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 1.9 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 4.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 3.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 3.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 3.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.1 | 0.4 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.6 | GO:0009931 | calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 4.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.0 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) very long-chain fatty acid-CoA ligase activity(GO:0031957) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 0.7 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 5.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 4.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.6 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 2.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.0 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 2.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 3.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.6 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.1 | 2.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 7.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.1 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.2 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.3 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 5.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 1.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 4.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 3.4 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 1.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 1.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 2.0 | GO:0015399 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) |
| 0.0 | 1.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 3.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.7 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.2 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |