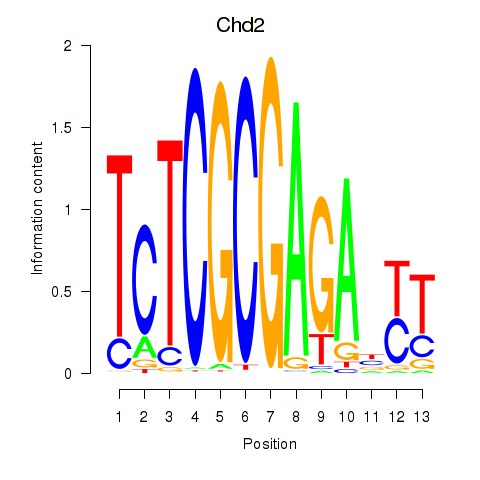
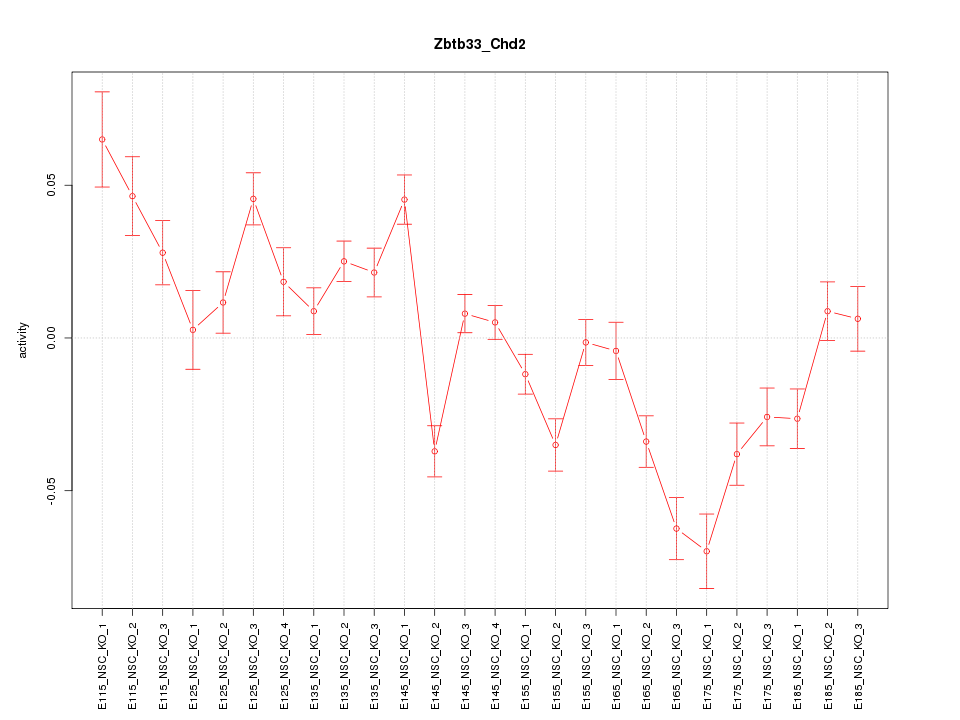
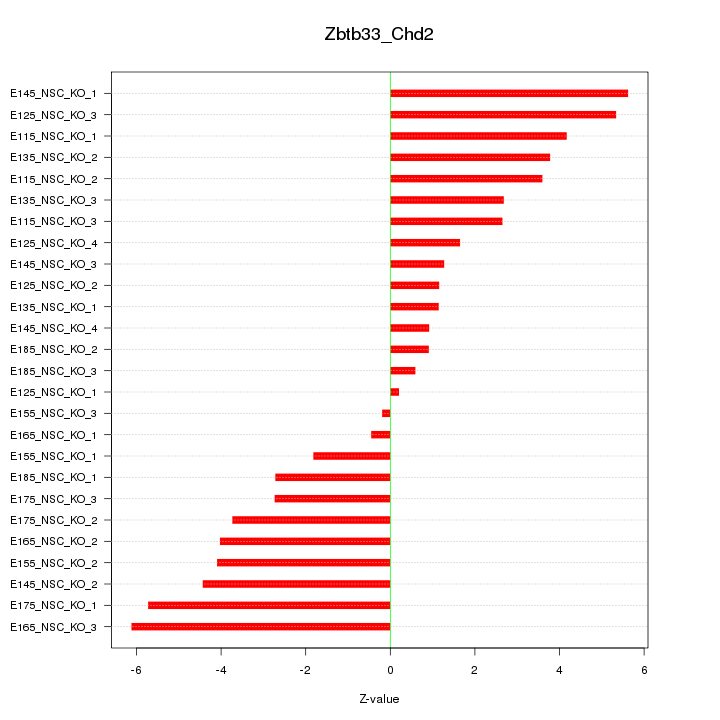
| 3.9 |
19.5 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 3.7 |
25.9 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 3.6 |
14.5 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 2.1 |
6.3 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 1.9 |
7.5 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 1.9 |
5.7 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 1.8 |
5.3 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 1.7 |
5.2 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 1.6 |
8.1 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 1.5 |
9.3 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 1.5 |
4.6 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.5 |
5.8 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 1.4 |
4.3 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.4 |
7.1 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 1.4 |
19.2 |
GO:0071459 |
protein localization to chromosome, centromeric region(GO:0071459) |
| 1.3 |
4.0 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 1.3 |
3.9 |
GO:1903722 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) regulation of centriole elongation(GO:1903722) |
| 1.3 |
12.5 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.2 |
1.2 |
GO:0000460 |
maturation of 5.8S rRNA(GO:0000460) |
| 1.1 |
8.8 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 1.1 |
5.5 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 1.1 |
3.3 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 1.1 |
3.2 |
GO:0034476 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 1.0 |
3.0 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.0 |
6.9 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 1.0 |
7.7 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.9 |
3.8 |
GO:0051593 |
response to folic acid(GO:0051593) |
| 0.9 |
4.6 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.9 |
4.3 |
GO:0003223 |
ventricular compact myocardium morphogenesis(GO:0003223) proprioception(GO:0019230) |
| 0.9 |
5.1 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.8 |
3.2 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.8 |
5.7 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.8 |
4.0 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.8 |
4.0 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.8 |
1.5 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 0.8 |
2.3 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.7 |
2.2 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.7 |
1.5 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.7 |
5.1 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.7 |
11.0 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.7 |
2.7 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.7 |
10.4 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.6 |
6.4 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.6 |
3.8 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.6 |
15.8 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) |
| 0.6 |
16.6 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.6 |
2.4 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.6 |
10.8 |
GO:0031297 |
replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.6 |
4.2 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.6 |
3.0 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.6 |
2.2 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.6 |
1.1 |
GO:0003169 |
coronary vein morphogenesis(GO:0003169) |
| 0.5 |
3.7 |
GO:0033567 |
DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.5 |
1.6 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.5 |
4.2 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.5 |
3.1 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.5 |
1.5 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.5 |
7.1 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.5 |
2.0 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.5 |
2.5 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.5 |
2.0 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.5 |
1.9 |
GO:0090069 |
regulation of ribosome biogenesis(GO:0090069) |
| 0.5 |
2.8 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.5 |
5.1 |
GO:0060340 |
positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.5 |
0.9 |
GO:0042505 |
tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.4 |
1.3 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 |
4.5 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.4 |
0.9 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.4 |
3.0 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 |
1.6 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.4 |
2.5 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.4 |
2.8 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 |
1.6 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 |
2.0 |
GO:1901838 |
positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.4 |
5.0 |
GO:1904851 |
positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.4 |
3.8 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.4 |
11.6 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 |
1.9 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.4 |
1.8 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 |
14.2 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.3 |
1.0 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 |
0.7 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.3 |
2.4 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.3 |
3.7 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 |
3.8 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.3 |
1.0 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.3 |
7.9 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.3 |
0.9 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.3 |
3.3 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.3 |
2.6 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 |
0.6 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.3 |
1.2 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.3 |
0.8 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.3 |
2.5 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.3 |
3.8 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.3 |
2.2 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 |
8.2 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.3 |
1.1 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.3 |
6.3 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.3 |
0.8 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.3 |
1.5 |
GO:0000055 |
ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 |
1.0 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.2 |
1.6 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.2 |
1.9 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 |
2.8 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.2 |
4.1 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 |
2.7 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 |
2.0 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.2 |
1.6 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 |
2.6 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 |
1.4 |
GO:0010804 |
negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 |
0.8 |
GO:1902255 |
membrane fission(GO:0090148) regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) regulation of membrane tubulation(GO:1903525) |
| 0.2 |
3.5 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.2 |
1.4 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.2 |
0.6 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 |
2.3 |
GO:0046349 |
amino sugar biosynthetic process(GO:0046349) |
| 0.2 |
1.1 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 |
2.4 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.2 |
0.6 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.2 |
2.2 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 |
0.9 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 |
0.5 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 |
12.8 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.2 |
0.7 |
GO:0046125 |
pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 |
0.7 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 |
0.5 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 |
4.0 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.2 |
0.8 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.2 |
3.1 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.2 |
1.8 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 |
2.4 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.2 |
0.9 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.2 |
1.4 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 |
10.2 |
GO:2000179 |
positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.2 |
3.4 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.2 |
2.1 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
0.9 |
GO:0051096 |
regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 |
1.0 |
GO:0030050 |
vesicle transport along actin filament(GO:0030050) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
1.1 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 |
1.4 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.1 |
1.7 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.1 |
3.6 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.1 |
0.7 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
14.7 |
GO:0031109 |
microtubule polymerization or depolymerization(GO:0031109) |
| 0.1 |
0.6 |
GO:1904690 |
adenosine to inosine editing(GO:0006382) regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 |
1.4 |
GO:0006517 |
protein deglycosylation(GO:0006517) |
| 0.1 |
0.7 |
GO:0036123 |
histone H3-K9 dimethylation(GO:0036123) |
| 0.1 |
4.6 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.1 |
0.3 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.1 |
0.3 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.1 |
3.1 |
GO:0035886 |
vascular smooth muscle cell differentiation(GO:0035886) |
| 0.1 |
0.9 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.1 |
2.2 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.1 |
1.2 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.4 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 |
0.2 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
3.2 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
1.8 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
0.5 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
1.5 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 |
1.6 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.1 |
0.8 |
GO:0006309 |
apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
0.1 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 |
2.2 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 |
1.4 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.1 |
0.9 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 |
0.5 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
0.2 |
GO:0035799 |
ureter maturation(GO:0035799) mast cell differentiation(GO:0060374) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 |
0.7 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
10.7 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 |
0.9 |
GO:0030238 |
male sex determination(GO:0030238) Leydig cell differentiation(GO:0033327) |
| 0.1 |
0.7 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
0.6 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.9 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.1 |
2.0 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.1 |
3.7 |
GO:0044255 |
cellular lipid metabolic process(GO:0044255) |
| 0.1 |
0.2 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
2.9 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 |
1.1 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.1 |
1.4 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.1 |
0.7 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.4 |
GO:0045723 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 |
0.7 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.1 |
6.3 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.1 |
1.4 |
GO:0018105 |
peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 |
1.8 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
2.1 |
GO:0032835 |
glomerulus development(GO:0032835) |
| 0.0 |
0.3 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
1.1 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.6 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
1.2 |
GO:0051567 |
histone H3-K9 methylation(GO:0051567) |
| 0.0 |
1.3 |
GO:0045860 |
positive regulation of protein kinase activity(GO:0045860) |
| 0.0 |
1.2 |
GO:0042060 |
wound healing(GO:0042060) |
| 0.0 |
0.5 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.7 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.7 |
GO:0010883 |
regulation of lipid storage(GO:0010883) |
| 0.0 |
10.3 |
GO:0006281 |
DNA repair(GO:0006281) |
| 0.0 |
1.0 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.9 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
0.2 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 |
0.3 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
2.1 |
GO:0016197 |
endosomal transport(GO:0016197) |
| 0.0 |
0.3 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.4 |
GO:0032366 |
intracellular sterol transport(GO:0032366) |
| 0.0 |
9.2 |
GO:0008380 |
RNA splicing(GO:0008380) |
| 0.0 |
0.5 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.2 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.0 |
0.1 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 |
1.0 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.6 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
0.3 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.1 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.0 |
1.5 |
GO:0001935 |
endothelial cell proliferation(GO:0001935) |
| 0.0 |
1.0 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
1.7 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
4.4 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
4.2 |
GO:0007517 |
muscle organ development(GO:0007517) |
| 0.0 |
0.2 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
1.0 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.4 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
3.8 |
GO:0007283 |
spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 |
1.0 |
GO:0048469 |
cell maturation(GO:0048469) |
| 0.0 |
2.3 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.1 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 |
0.3 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.4 |
GO:0032204 |
regulation of telomere maintenance(GO:0032204) |
| 0.0 |
0.4 |
GO:0032436 |
positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |