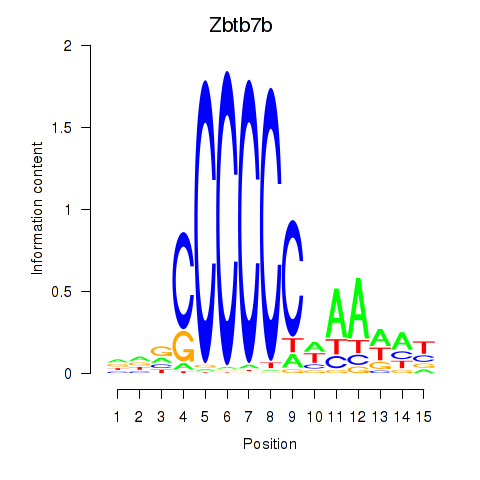
| 1.1 |
4.4 |
GO:2000054 |
negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.1 |
3.2 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 1.0 |
8.6 |
GO:1902715 |
positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.7 |
3.4 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.6 |
2.3 |
GO:0046874 |
quinolinate metabolic process(GO:0046874) |
| 0.5 |
1.6 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 |
3.3 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.5 |
1.9 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 |
1.7 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.4 |
1.7 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.4 |
1.1 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.4 |
1.1 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 |
3.5 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 |
2.6 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.2 |
1.7 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.2 |
0.7 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.2 |
1.7 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 |
0.7 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.2 |
0.6 |
GO:1902071 |
regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 |
1.6 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.2 |
3.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 |
1.1 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 |
1.0 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.2 |
1.1 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 |
0.8 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
1.6 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
0.8 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.5 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
2.4 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.1 |
1.0 |
GO:0036123 |
histone H3-K9 dimethylation(GO:0036123) |
| 0.1 |
0.3 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
1.7 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.1 |
0.5 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.5 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 |
1.1 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 0.1 |
0.8 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.2 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.7 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 |
0.7 |
GO:0060340 |
positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 |
0.3 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.1 |
0.2 |
GO:0002925 |
positive regulation of chronic inflammatory response(GO:0002678) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 |
0.4 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 |
1.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.2 |
GO:1902915 |
negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.2 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.2 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 |
2.2 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 |
0.6 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) negative regulation of protein folding(GO:1903333) |
| 0.1 |
3.9 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.1 |
0.1 |
GO:0044340 |
canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 |
0.9 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.9 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.5 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.0 |
0.6 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 |
0.6 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
0.3 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 |
0.2 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.2 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 |
0.2 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
1.1 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.1 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 |
1.4 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
1.1 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.1 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.3 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.7 |
GO:0031065 |
positive regulation of histone deacetylation(GO:0031065) |
| 0.0 |
1.6 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 |
0.1 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.0 |
2.0 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 |
0.2 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.2 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 |
0.7 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.7 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.6 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.4 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
0.4 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.4 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
1.2 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.3 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.0 |
GO:0071455 |
cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 |
0.3 |
GO:0060219 |
camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 |
0.3 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.2 |
GO:0071357 |
type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 |
0.3 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
0.4 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.5 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
2.0 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.5 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.6 |
GO:0002209 |
behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |