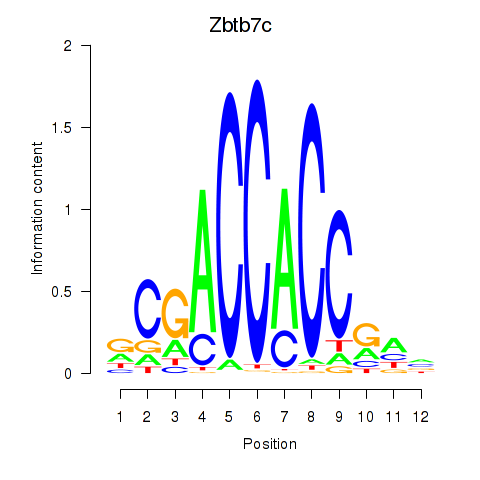
| 1.7 |
7.0 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.0 |
15.6 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.6 |
1.7 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.4 |
2.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 |
5.6 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 |
0.9 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.3 |
1.3 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.2 |
1.0 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.2 |
1.9 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.2 |
4.1 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 |
2.0 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 |
0.6 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 |
1.3 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.2 |
0.5 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) trachea cartilage morphogenesis(GO:0060535) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.1 |
1.8 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.7 |
GO:0090467 |
lysine transport(GO:0015819) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 |
1.8 |
GO:0032438 |
melanosome organization(GO:0032438) endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 |
0.4 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 |
0.5 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
1.3 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.1 |
1.6 |
GO:0070445 |
radial glial cell differentiation(GO:0060019) oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 |
0.5 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 |
1.4 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.1 |
0.8 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
1.1 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 |
1.7 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 |
4.9 |
GO:0015807 |
L-amino acid transport(GO:0015807) |
| 0.1 |
0.2 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.1 |
0.8 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
4.2 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.1 |
3.2 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.0 |
0.2 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
1.7 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.7 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
1.7 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.2 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
8.9 |
GO:0007411 |
axon guidance(GO:0007411) |
| 0.0 |
0.1 |
GO:0014022 |
neural plate morphogenesis(GO:0001839) neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 |
4.7 |
GO:0071805 |
potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.8 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.7 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.4 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.7 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
4.0 |
GO:0007409 |
axonogenesis(GO:0007409) |
| 0.0 |
0.4 |
GO:0010955 |
negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 |
0.8 |
GO:0006919 |
activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 |
0.2 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
1.8 |
GO:0051291 |
protein heterooligomerization(GO:0051291) |
| 0.0 |
0.2 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.3 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |