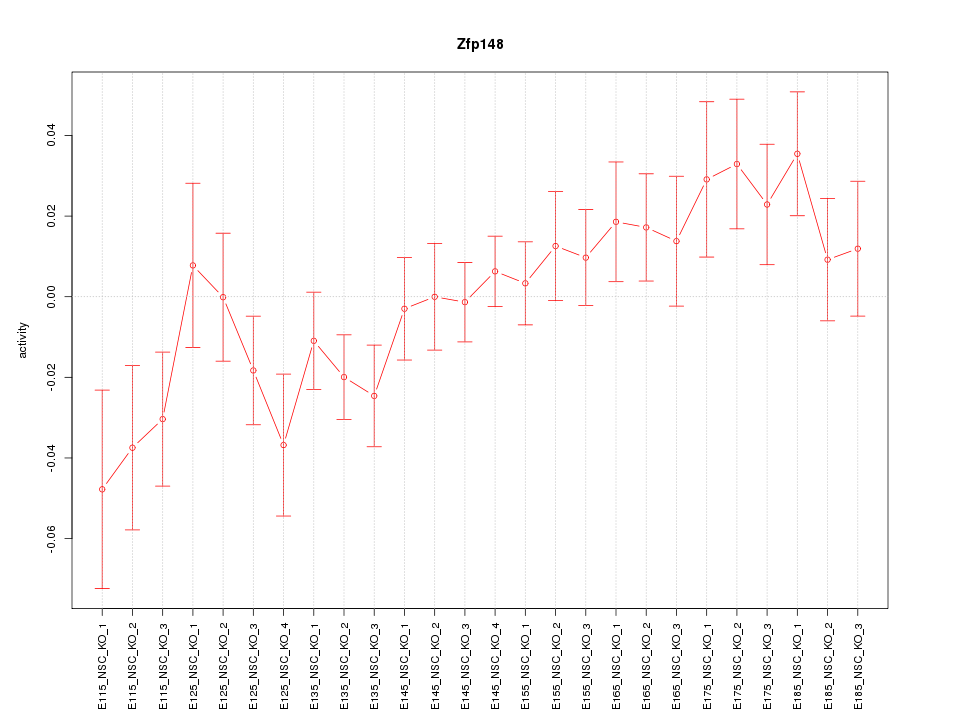
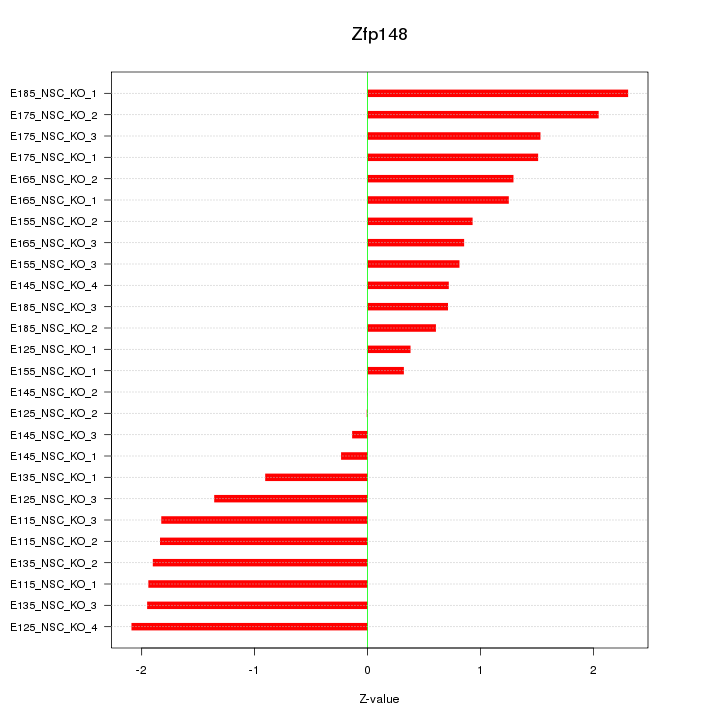
Motif ID: Zfp148
Z-value: 1.334
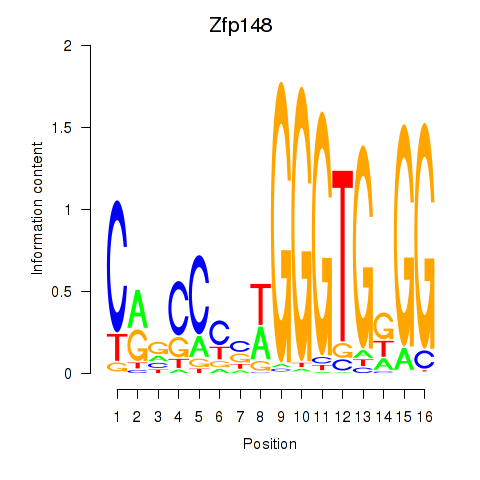
Transcription factors associated with Zfp148:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp148 | ENSMUSG00000022811.10 | Zfp148 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp148 | mm10_v2_chr16_+_33380765_33380787 | 0.14 | 4.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.8 | 10.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.5 | 9.0 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 1.1 | 3.4 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.0 | 4.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 1.0 | 2.9 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.9 | 2.8 | GO:0042939 | renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 | 2.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.8 | 2.5 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.7 | 2.2 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.6 | 1.9 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.6 | 3.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.6 | 2.2 | GO:0097494 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of vesicle size(GO:0097494) |
| 0.5 | 1.6 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.5 | 2.1 | GO:0061428 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.5 | 2.6 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.5 | 1.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.5 | 4.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.5 | 2.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.5 | 1.9 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.5 | 5.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.5 | 2.3 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.4 | 3.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 | 1.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 1.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.4 | 1.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.4 | 1.9 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.4 | 1.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.4 | 1.8 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.4 | 1.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 1.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.3 | 3.8 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.3 | 3.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.3 | 1.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 24.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 2.6 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 2.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 1.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 0.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 4.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 0.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.9 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 2.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 1.8 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 1.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 | 0.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 2.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 0.5 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 4.4 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.6 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.9 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 1.3 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.5 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 2.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 3.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.2 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.1 | 1.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 1.6 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.3 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 1.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.1 | 4.0 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.1 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.7 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 3.8 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.1 | 7.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.2 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.4 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 1.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 4.8 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 7.2 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 2.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 1.3 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 1.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 1.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.2 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 3.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 1.7 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.5 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 1.9 | GO:0051297 | centrosome organization(GO:0051297) |
| 0.0 | 2.1 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 2.8 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 1.3 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.6 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 1.9 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.5 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 2.2 | 9.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.0 | 1.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 2.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.6 | 3.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 3.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.6 | 2.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 3.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) dendritic spine neck(GO:0044326) |
| 0.3 | 2.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 2.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 1.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 3.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 4.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 1.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 3.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 18.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 7.9 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 1.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 16.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 10.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 8.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 6.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 5.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.8 | GO:0070469 | respiratory chain(GO:0070469) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.5 | 16.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.0 | 3.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.9 | 7.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.6 | 5.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 2.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.5 | 2.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.5 | 3.4 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 3.8 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.5 | 1.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 1.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 1.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 0.4 | 1.9 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 1.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 4.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 2.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 2.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 2.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 3.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 1.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 5.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 2.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 2.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 2.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 7.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 3.1 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 10.0 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.1 | 0.4 | GO:0019798 | procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 8.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) very long-chain fatty acid-CoA ligase activity(GO:0031957) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 1.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 4.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.8 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 3.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 2.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.7 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 1.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 3.7 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |