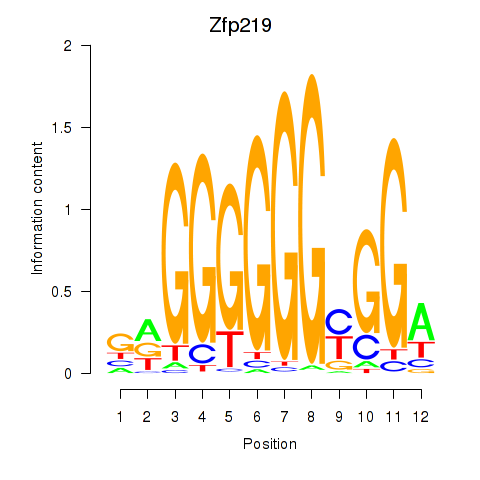
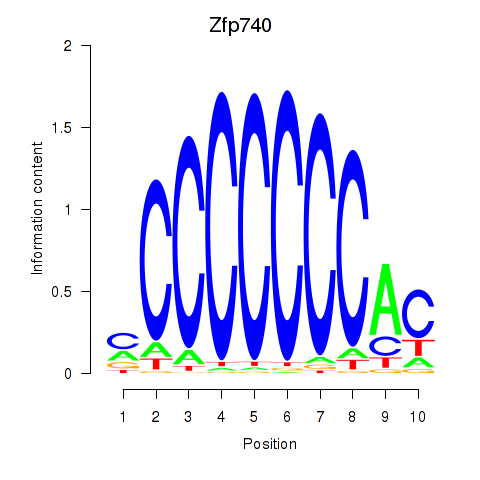
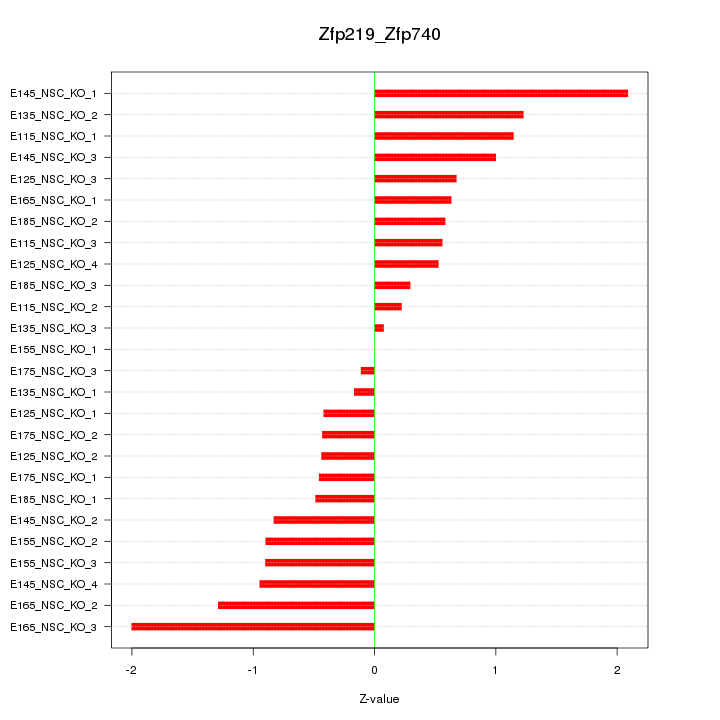
| 0.8 |
2.3 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 0.7 |
3.4 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.6 |
1.7 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.5 |
1.6 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.5 |
5.3 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.5 |
2.1 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.5 |
2.0 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.5 |
1.5 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.5 |
6.8 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.4 |
1.7 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.4 |
2.8 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.4 |
0.8 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.4 |
1.1 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.4 |
1.8 |
GO:1900109 |
regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.3 |
4.4 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.3 |
1.3 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 |
2.9 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.3 |
0.9 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.3 |
2.4 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.3 |
1.8 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 |
0.6 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.3 |
1.1 |
GO:0051305 |
chromosome movement towards spindle pole(GO:0051305) |
| 0.3 |
2.0 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.3 |
0.6 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 |
0.8 |
GO:0021557 |
oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 |
0.8 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.3 |
1.3 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.2 |
0.5 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.2 |
0.7 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 |
0.9 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 |
0.7 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.2 |
0.6 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 |
0.8 |
GO:0035878 |
nail development(GO:0035878) |
| 0.2 |
0.6 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 |
4.0 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.2 |
2.1 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 |
0.6 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.2 |
0.7 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 |
0.7 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 |
1.1 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 |
0.7 |
GO:0007223 |
muscular septum morphogenesis(GO:0003150) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 |
0.9 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.2 |
1.4 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.2 |
0.7 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.2 |
1.2 |
GO:0070814 |
hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.2 |
0.5 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.2 |
1.2 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 |
1.8 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 |
0.6 |
GO:0097374 |
sensory neuron axon guidance(GO:0097374) |
| 0.2 |
0.6 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.2 |
0.9 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.2 |
0.6 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.2 |
0.8 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.4 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.1 |
1.2 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 |
0.7 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 |
0.8 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.4 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
1.7 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.9 |
GO:0051461 |
positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
2.1 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.1 |
0.4 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
0.8 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.1 |
0.5 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 |
0.5 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.1 |
1.2 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 |
0.3 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 |
0.3 |
GO:0060129 |
thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 |
0.3 |
GO:0070602 |
regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 |
1.3 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
0.3 |
GO:0003192 |
mitral valve formation(GO:0003192) |
| 0.1 |
0.8 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.1 |
0.4 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.1 |
0.7 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.4 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.1 |
0.3 |
GO:1904683 |
regulation of metalloendopeptidase activity(GO:1904683) negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
2.1 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.1 |
0.5 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.1 |
0.7 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 |
0.3 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 |
0.5 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 |
0.3 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 |
0.6 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.4 |
GO:0071105 |
negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 |
0.8 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.4 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 |
0.3 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.4 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
0.3 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
2.0 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 |
0.7 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.2 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 |
0.4 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 |
0.6 |
GO:0090188 |
negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.1 |
0.2 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 |
0.4 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
1.2 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.4 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.7 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
0.3 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 |
1.2 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.1 |
0.1 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 |
0.3 |
GO:1901163 |
trophoblast cell migration(GO:0061450) regulation of trophoblast cell migration(GO:1901163) |
| 0.1 |
3.3 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.1 |
0.3 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.1 |
0.2 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 |
0.7 |
GO:0071578 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) zinc II ion transmembrane import(GO:0071578) |
| 0.1 |
0.5 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
0.7 |
GO:0030853 |
negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 |
0.7 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.1 |
0.1 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.1 |
0.3 |
GO:0097393 |
negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.1 |
0.2 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.1 |
0.4 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 |
0.7 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.3 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
1.0 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.7 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 |
0.2 |
GO:0060982 |
coronary artery morphogenesis(GO:0060982) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
0.1 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 |
0.2 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.1 |
0.4 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 |
0.4 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
1.3 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.1 |
1.3 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 |
0.4 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.3 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.1 |
GO:0033092 |
positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 |
0.5 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.4 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 |
0.5 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.2 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.6 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.2 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.0 |
1.3 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.6 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 0.0 |
0.4 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 |
1.4 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.4 |
GO:1903069 |
regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 |
0.1 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.2 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.4 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 |
0.5 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.3 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 |
2.1 |
GO:0051384 |
response to glucocorticoid(GO:0051384) |
| 0.0 |
0.7 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
1.0 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.7 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.9 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 |
0.2 |
GO:0045072 |
regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
2.2 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.7 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.1 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.2 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 |
0.6 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.0 |
0.5 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.4 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.3 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
1.0 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.8 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.4 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.2 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.0 |
0.1 |
GO:0060431 |
primary lung bud formation(GO:0060431) |
| 0.0 |
0.4 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.3 |
GO:0055090 |
acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 |
0.5 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.6 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.0 |
0.1 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.0 |
0.7 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.4 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.3 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.0 |
0.3 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
1.3 |
GO:0045638 |
negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 |
0.4 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.4 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.2 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.3 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.2 |
GO:0071455 |
cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
1.1 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.6 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.9 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.3 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.8 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.8 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.3 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
2.0 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.6 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.6 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.2 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.1 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 |
0.1 |
GO:0043046 |
DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 |
0.1 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.2 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.4 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.0 |
0.1 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
1.2 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.1 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.6 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.1 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.0 |
GO:0001787 |
natural killer cell proliferation(GO:0001787) |
| 0.0 |
0.1 |
GO:1903975 |
regulation of glial cell migration(GO:1903975) |
| 0.0 |
0.3 |
GO:0008016 |
regulation of heart contraction(GO:0008016) |
| 0.0 |
0.2 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.2 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.6 |
GO:0030326 |
embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 |
0.2 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.0 |
0.6 |
GO:0045727 |
positive regulation of translation(GO:0045727) |