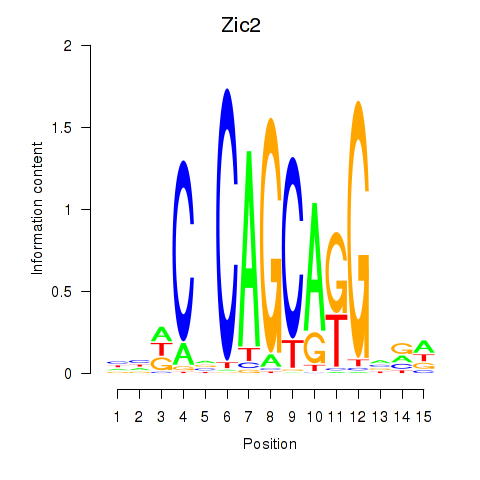
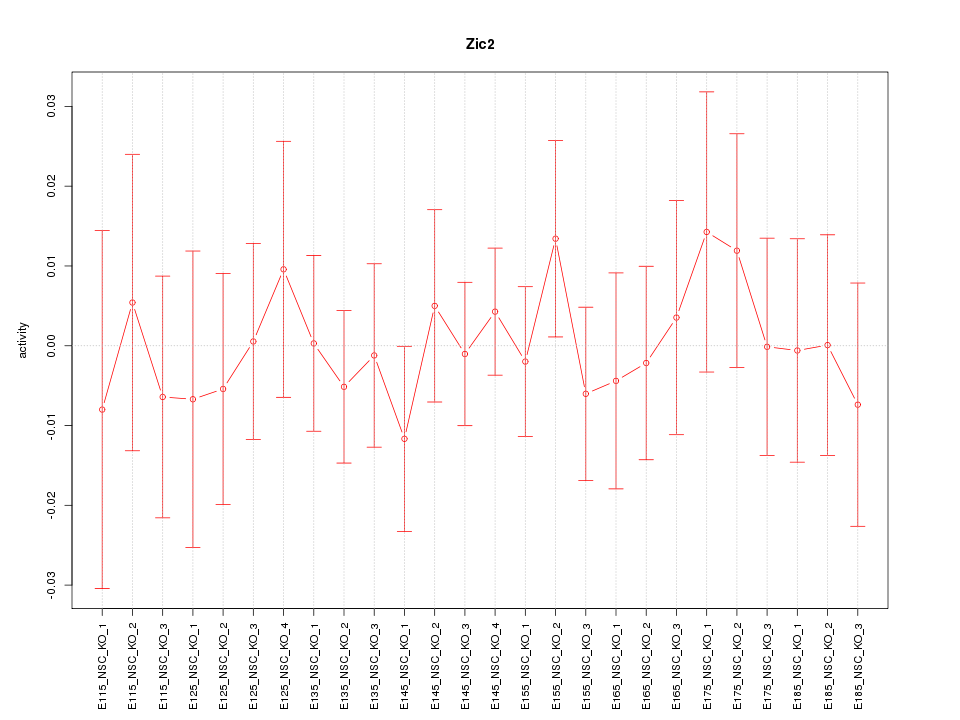
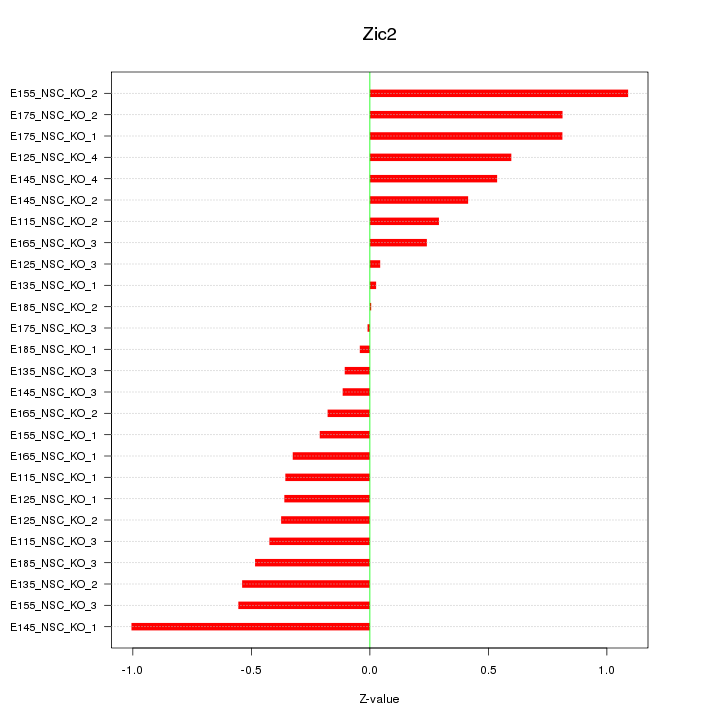
| 0.7 |
2.2 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.3 |
1.1 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 |
2.0 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.2 |
0.5 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.2 |
0.5 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.2 |
0.7 |
GO:1903802 |
positive regulation of arachidonic acid secretion(GO:0090238) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.2 |
0.8 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.4 |
GO:0035441 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) cell migration involved in vasculogenesis(GO:0035441) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) ganglion morphogenesis(GO:0061552) positive regulation of retinal ganglion cell axon guidance(GO:1902336) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.1 |
0.7 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.1 |
0.4 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.1 |
0.3 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 0.1 |
0.3 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 |
0.8 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.7 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.3 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
0.3 |
GO:2000551 |
regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.1 |
0.2 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.1 |
0.4 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.4 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 |
0.2 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.1 |
0.5 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.3 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 |
0.2 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.1 |
0.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.4 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
0.3 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.3 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.1 |
0.2 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 |
0.7 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.2 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) cellular response to copper ion(GO:0071280) |
| 0.1 |
0.2 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.1 |
0.3 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.6 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
0.1 |
GO:0015819 |
lysine transport(GO:0015819) |
| 0.1 |
0.3 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.2 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.0 |
0.1 |
GO:0010898 |
positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 |
0.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.2 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 |
0.2 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 |
0.1 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.0 |
0.4 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
1.0 |
GO:0010043 |
response to zinc ion(GO:0010043) |
| 0.0 |
0.1 |
GO:0046959 |
habituation(GO:0046959) |
| 0.0 |
0.3 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.0 |
0.2 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.4 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.2 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.1 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 |
0.1 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 |
0.5 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.1 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.1 |
GO:0097491 |
trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 |
0.2 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 |
0.1 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 |
0.1 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.0 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.1 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 |
0.2 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 |
0.3 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.1 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.4 |
GO:0006047 |
UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 |
0.4 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.1 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 |
0.1 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.0 |
0.1 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 |
0.3 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.2 |
GO:0030388 |
fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.2 |
GO:0003093 |
regulation of glomerular filtration(GO:0003093) |
| 0.0 |
0.2 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:0046078 |
pyrimidine deoxyribonucleotide catabolic process(GO:0009223) dUMP metabolic process(GO:0046078) |
| 0.0 |
0.1 |
GO:0070560 |
thrombin receptor signaling pathway(GO:0070493) protein secretion by platelet(GO:0070560) |
| 0.0 |
0.1 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.2 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.2 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 |
0.6 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.2 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 |
0.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.7 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
0.1 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) male mating behavior(GO:0060179) organism emergence from protective structure(GO:0071684) |
| 0.0 |
0.1 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 |
0.3 |
GO:0032026 |
response to magnesium ion(GO:0032026) |
| 0.0 |
0.1 |
GO:0035509 |
negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 |
0.2 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.3 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.1 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.1 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 |
0.1 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.2 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.2 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.0 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 |
0.5 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.2 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.0 |
0.0 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) regulation of vascular wound healing(GO:0061043) |
| 0.0 |
0.0 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 |
0.1 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 |
0.3 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 |
0.0 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.0 |
0.5 |
GO:2000649 |
regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.2 |
GO:2000398 |
regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 |
0.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.0 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.1 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.0 |
0.2 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.1 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.5 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.9 |
GO:0030279 |
negative regulation of ossification(GO:0030279) |
| 0.0 |
0.1 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.3 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.6 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |