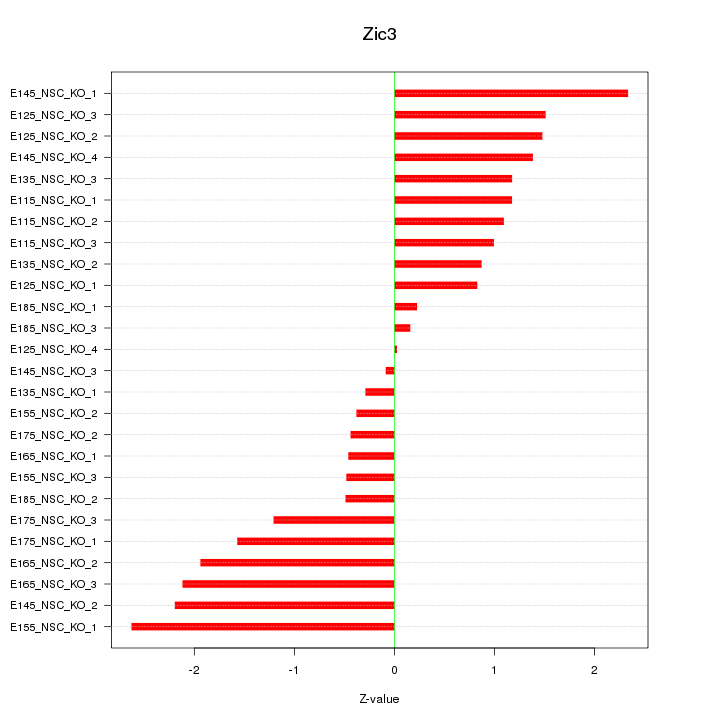
Motif ID: Zic3
Z-value: 1.291

Transcription factors associated with Zic3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zic3 | ENSMUSG00000067860.5 | Zic3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic3 | mm10_v2_chrX_+_58030999_58031018 | 0.77 | 4.4e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0097402 | neuroblast migration(GO:0097402) |
| 1.0 | 5.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.8 | 2.5 | GO:0046671 | negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.8 | 3.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.8 | 8.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.7 | 2.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.7 | 3.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.7 | 9.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.6 | 3.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 2.4 | GO:0035624 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.5 | 1.6 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.5 | 1.5 | GO:0070103 | interleukin-4-mediated signaling pathway(GO:0035771) tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.5 | 4.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.5 | 3.9 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.5 | 1.9 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.4 | 1.8 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.4 | 1.8 | GO:0060032 | notochord regression(GO:0060032) |
| 0.4 | 1.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.4 | 1.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.4 | 1.7 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.4 | 1.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) glutamate secretion, neurotransmission(GO:0061535) |
| 0.4 | 1.1 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.4 | 1.1 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 2.9 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 1.1 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.4 | 1.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 2.8 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 1.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.3 | 1.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.3 | 3.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 1.6 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 | 5.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.3 | 2.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 2.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.3 | 2.5 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 | 1.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.3 | 0.3 | GO:2000224 | testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.3 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 3.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.3 | 0.8 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 0.7 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 0.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 0.7 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.2 | 1.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 0.4 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.2 | 1.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.6 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.2 | 3.0 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 0.6 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 | 0.8 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 1.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 3.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 0.7 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 2.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 1.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.5 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 5.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.8 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 0.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.5 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 1.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.4 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.9 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.6 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.3 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.1 | 1.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.6 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.0 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.7 | GO:0070474 | regulation of uterine smooth muscle contraction(GO:0070472) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 1.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.6 | GO:2000301 | myeloid progenitor cell differentiation(GO:0002318) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.0 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.1 | 1.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.2 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 1.6 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.7 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 1.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.5 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 1.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.3 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.7 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.5 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 1.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.9 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.5 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.6 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 1.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.0 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.8 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 1.3 | GO:0006096 | gluconeogenesis(GO:0006094) glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.4 | GO:2000816 | negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.5 | 3.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 1.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.4 | 3.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 1.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 1.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 3.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 5.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 0.7 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 2.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 0.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 2.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 4.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.9 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 2.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.5 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.6 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.6 | 2.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.6 | 2.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.6 | 3.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 2.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 5.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 1.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 4.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 2.8 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.3 | 2.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 2.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 1.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.3 | 3.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 0.7 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 1.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 1.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 1.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 0.8 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.1 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.2 | 1.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 3.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 1.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 4.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.5 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.6 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 0.3 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.1 | 1.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.8 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.1 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.9 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 3.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.1 | 1.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.2 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.1 | 1.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.6 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 4.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 4.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 8.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 1.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.9 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.8 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 5.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.7 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |