Project
12D miR HR13_24
Navigation
Downloads
Results for Rela_Rel_Nfkb1
Z-value: 2.14
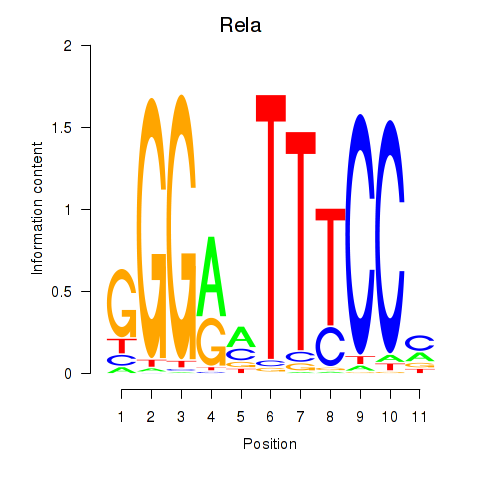
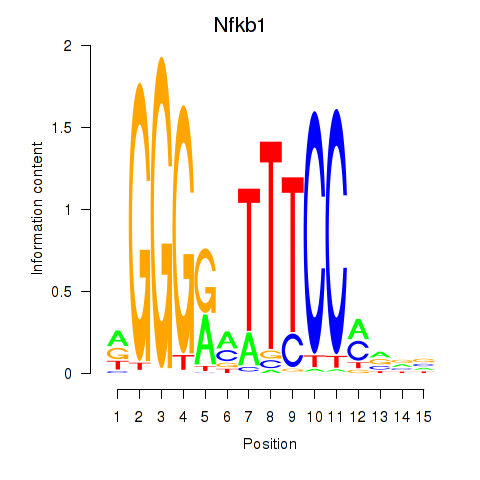
Transcription factors associated with Rela_Rel_Nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rela
|
ENSMUSG00000024927.7 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) |
|
Rel
|
ENSMUSG00000020275.8 | reticuloendotheliosis oncogene |
|
Nfkb1
|
ENSMUSG00000028163.11 | nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb1 | mm10_v2_chr3_-_135691512_135691564 | -0.81 | 2.2e-03 | Click! |
| Rela | mm10_v2_chr19_+_5637475_5637486 | -0.70 | 1.6e-02 | Click! |
| Rel | mm10_v2_chr11_-_23770953_23771028 | -0.64 | 3.3e-02 | Click! |
Activity profile of Rela_Rel_Nfkb1 motif
Sorted Z-values of Rela_Rel_Nfkb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_165234689 | 4.64 |
ENSMUST00000065438.6
|
Cdh22
|
cadherin 22 |
| chr5_+_90891234 | 3.80 |
ENSMUST00000031327.8
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr5_-_92348871 | 3.44 |
ENSMUST00000038816.6
ENSMUST00000118006.1 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr5_+_90759299 | 3.24 |
ENSMUST00000031318.4
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr4_+_150927918 | 3.21 |
ENSMUST00000139826.1
ENSMUST00000116257.1 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr15_-_31367527 | 3.20 |
ENSMUST00000076942.4
ENSMUST00000123325.1 ENSMUST00000110410.2 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr7_-_44815658 | 3.06 |
ENSMUST00000107893.1
|
Atf5
|
activating transcription factor 5 |
| chr9_-_7873157 | 2.85 |
ENSMUST00000159323.1
ENSMUST00000115673.2 |
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr9_-_7873016 | 2.67 |
ENSMUST00000013949.8
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr13_+_112464070 | 2.65 |
ENSMUST00000183663.1
ENSMUST00000184311.1 ENSMUST00000183886.1 |
Il6st
|
interleukin 6 signal transducer |
| chr2_-_32387760 | 2.46 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr1_+_150100093 | 2.44 |
ENSMUST00000035065.7
|
Ptgs2
|
prostaglandin-endoperoxide synthase 2 |
| chr19_+_6341121 | 2.38 |
ENSMUST00000025897.6
ENSMUST00000130382.1 |
Map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr1_-_135258449 | 2.27 |
ENSMUST00000003135.7
|
Elf3
|
E74-like factor 3 |
| chr9_-_7872983 | 2.23 |
ENSMUST00000115672.1
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr18_-_4352944 | 2.22 |
ENSMUST00000025078.2
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr14_-_30607808 | 2.17 |
ENSMUST00000112207.1
ENSMUST00000112206.1 ENSMUST00000112202.1 ENSMUST00000112203.1 |
Prkcd
|
protein kinase C, delta |
| chr19_+_46304709 | 2.10 |
ENSMUST00000073116.5
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr19_-_58454435 | 2.10 |
ENSMUST00000169850.1
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr19_-_58454580 | 2.09 |
ENSMUST00000129100.1
ENSMUST00000123957.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr4_+_115088708 | 2.09 |
ENSMUST00000171877.1
ENSMUST00000177647.1 ENSMUST00000106548.2 ENSMUST00000030488.2 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr17_-_34287770 | 1.99 |
ENSMUST00000174751.1
ENSMUST00000040655.6 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr2_-_103303179 | 1.99 |
ENSMUST00000090475.3
|
Ehf
|
ets homologous factor |
| chr2_-_5063996 | 1.95 |
ENSMUST00000114996.1
|
Optn
|
optineurin |
| chr2_-_103303158 | 1.95 |
ENSMUST00000111176.2
|
Ehf
|
ets homologous factor |
| chr11_+_100860447 | 1.93 |
ENSMUST00000107357.2
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr11_-_109298066 | 1.92 |
ENSMUST00000106706.1
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr13_+_18948344 | 1.90 |
ENSMUST00000003345.7
|
Amph
|
amphiphysin |
| chr7_+_28766747 | 1.89 |
ENSMUST00000170068.1
ENSMUST00000072965.4 |
Sirt2
|
sirtuin 2 |
| chrX_+_164438039 | 1.88 |
ENSMUST00000033755.5
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr11_+_93098404 | 1.87 |
ENSMUST00000107859.1
ENSMUST00000042943.6 ENSMUST00000107861.1 ENSMUST00000107858.2 |
Car10
|
carbonic anhydrase 10 |
| chr9_+_22188137 | 1.84 |
ENSMUST00000178901.1
|
Zfp872
|
zinc finger protein 872 |
| chr17_+_35979851 | 1.82 |
ENSMUST00000087200.3
|
Gnl1
|
guanine nucleotide binding protein-like 1 |
| chr2_+_144556229 | 1.80 |
ENSMUST00000143573.1
ENSMUST00000028916.8 ENSMUST00000155258.1 |
Sec23b
|
SEC23B (S. cerevisiae) |
| chr3_-_83841767 | 1.79 |
ENSMUST00000029623.9
|
Tlr2
|
toll-like receptor 2 |
| chr2_+_144556306 | 1.76 |
ENSMUST00000155876.1
ENSMUST00000149697.1 |
Sec23b
|
SEC23B (S. cerevisiae) |
| chrX_+_164506320 | 1.76 |
ENSMUST00000033756.2
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr2_-_5063932 | 1.76 |
ENSMUST00000027986.4
|
Optn
|
optineurin |
| chr18_-_20059478 | 1.67 |
ENSMUST00000075214.2
ENSMUST00000039247.4 |
Dsc2
|
desmocollin 2 |
| chr7_+_99535439 | 1.65 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chr9_+_46998931 | 1.65 |
ENSMUST00000178065.1
|
Gm4791
|
predicted gene 4791 |
| chr18_+_60803838 | 1.62 |
ENSMUST00000050487.8
ENSMUST00000097563.2 ENSMUST00000167610.1 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr17_+_33919332 | 1.61 |
ENSMUST00000025161.7
|
Tapbp
|
TAP binding protein |
| chr17_+_24878724 | 1.61 |
ENSMUST00000050714.6
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr2_-_152830615 | 1.58 |
ENSMUST00000146380.1
ENSMUST00000134902.1 ENSMUST00000134357.1 ENSMUST00000109820.3 |
Bcl2l1
|
BCL2-like 1 |
| chrX_+_103321398 | 1.52 |
ENSMUST00000033689.2
|
Cdx4
|
caudal type homeobox 4 |
| chr1_+_134193432 | 1.50 |
ENSMUST00000038445.6
|
Mybph
|
myosin binding protein H |
| chr11_-_109298121 | 1.46 |
ENSMUST00000020920.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr9_-_14614949 | 1.46 |
ENSMUST00000013220.6
ENSMUST00000160770.1 |
Amotl1
|
angiomotin-like 1 |
| chr11_-_106159902 | 1.43 |
ENSMUST00000064545.4
|
Limd2
|
LIM domain containing 2 |
| chr11_+_100860326 | 1.43 |
ENSMUST00000138083.1
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chrX_+_7657260 | 1.41 |
ENSMUST00000033485.7
|
Prickle3
|
prickle homolog 3 (Drosophila) |
| chr8_-_25101985 | 1.39 |
ENSMUST00000128715.1
ENSMUST00000064883.6 |
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr2_-_84743655 | 1.38 |
ENSMUST00000181711.1
|
Gm19426
|
predicted gene, 19426 |
| chr6_-_5256226 | 1.38 |
ENSMUST00000125686.1
ENSMUST00000031773.2 |
Pon3
|
paraoxonase 3 |
| chr19_-_58455161 | 1.38 |
ENSMUST00000135730.1
ENSMUST00000152507.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr10_-_19015347 | 1.36 |
ENSMUST00000019997.4
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_99626936 | 1.36 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chr4_+_100776664 | 1.35 |
ENSMUST00000030257.8
ENSMUST00000097955.2 |
Cachd1
|
cache domain containing 1 |
| chr7_+_75455534 | 1.31 |
ENSMUST00000147005.1
ENSMUST00000166315.1 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr12_-_55492587 | 1.29 |
ENSMUST00000021413.7
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr7_+_99535652 | 1.29 |
ENSMUST00000032995.8
ENSMUST00000162404.1 |
Arrb1
|
arrestin, beta 1 |
| chr5_-_105293699 | 1.29 |
ENSMUST00000050011.8
|
Gbp6
|
guanylate binding protein 6 |
| chr4_-_42168603 | 1.27 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr14_+_80000292 | 1.27 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr2_+_143546144 | 1.27 |
ENSMUST00000028905.9
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr8_+_84415348 | 1.19 |
ENSMUST00000121390.1
ENSMUST00000122053.1 |
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr12_+_79029150 | 1.16 |
ENSMUST00000039928.5
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr18_+_37473538 | 1.15 |
ENSMUST00000050034.1
|
Pcdhb15
|
protocadherin beta 15 |
| chr6_+_48841633 | 1.15 |
ENSMUST00000168406.1
|
Tmem176a
|
transmembrane protein 176A |
| chr17_+_55952623 | 1.15 |
ENSMUST00000003274.6
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr5_+_66968416 | 1.11 |
ENSMUST00000038188.7
|
Limch1
|
LIM and calponin homology domains 1 |
| chr10_-_127751707 | 1.11 |
ENSMUST00000079692.5
|
Gpr182
|
G protein-coupled receptor 182 |
| chr11_+_100859492 | 1.09 |
ENSMUST00000107356.1
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr2_-_60881360 | 1.09 |
ENSMUST00000164147.1
ENSMUST00000112509.1 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr2_-_164443177 | 1.08 |
ENSMUST00000017153.3
|
Sdc4
|
syndecan 4 |
| chr5_+_64970069 | 1.08 |
ENSMUST00000031080.8
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr4_-_125127817 | 1.05 |
ENSMUST00000036188.7
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr2_+_120567687 | 1.02 |
ENSMUST00000028743.3
ENSMUST00000116437.1 ENSMUST00000153580.1 ENSMUST00000142278.1 |
Snap23
|
synaptosomal-associated protein 23 |
| chr11_-_47379405 | 1.01 |
ENSMUST00000077221.5
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr6_+_48841476 | 1.01 |
ENSMUST00000101426.4
|
Tmem176a
|
transmembrane protein 176A |
| chr18_+_84851338 | 1.01 |
ENSMUST00000160180.1
|
Cyb5
|
cytochrome b-5 |
| chr11_-_54956047 | 1.01 |
ENSMUST00000155316.1
ENSMUST00000108889.3 ENSMUST00000126703.1 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr3_+_28263205 | 1.00 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr14_+_41131777 | 1.00 |
ENSMUST00000022314.3
ENSMUST00000170719.1 |
Sftpa1
|
surfactant associated protein A1 |
| chr3_-_102936093 | 1.00 |
ENSMUST00000029448.6
|
Sycp1
|
synaptonemal complex protein 1 |
| chrX_-_162565514 | 1.00 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr17_+_26933070 | 0.99 |
ENSMUST00000073724.5
|
Phf1
|
PHD finger protein 1 |
| chr2_-_157566319 | 0.99 |
ENSMUST00000109528.2
ENSMUST00000088494.2 |
Blcap
|
bladder cancer associated protein homolog (human) |
| chr11_-_109298090 | 0.98 |
ENSMUST00000106704.2
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr2_+_120567652 | 0.96 |
ENSMUST00000110711.2
|
Snap23
|
synaptosomal-associated protein 23 |
| chr11_+_87664274 | 0.96 |
ENSMUST00000092800.5
|
Rnf43
|
ring finger protein 43 |
| chr3_+_122729158 | 0.96 |
ENSMUST00000066728.5
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr1_-_136230289 | 0.96 |
ENSMUST00000150163.1
ENSMUST00000144464.1 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr11_+_87664549 | 0.94 |
ENSMUST00000121782.2
|
Rnf43
|
ring finger protein 43 |
| chr9_+_54698859 | 0.94 |
ENSMUST00000120452.1
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr1_+_135132693 | 0.93 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr4_+_102589687 | 0.93 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr11_+_53519725 | 0.92 |
ENSMUST00000108987.1
ENSMUST00000121334.1 ENSMUST00000117061.1 |
Sept8
|
septin 8 |
| chr8_-_25101734 | 0.92 |
ENSMUST00000098866.4
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr10_-_95415484 | 0.92 |
ENSMUST00000172070.1
ENSMUST00000150432.1 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr14_-_30626196 | 0.91 |
ENSMUST00000112210.3
ENSMUST00000112211.2 ENSMUST00000112208.1 |
Prkcd
|
protein kinase C, delta |
| chr13_+_33964659 | 0.90 |
ENSMUST00000021843.5
ENSMUST00000058978.7 |
Nqo2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr12_-_80132802 | 0.89 |
ENSMUST00000180643.1
|
2310015A10Rik
|
RIKEN cDNA 2310015A10 gene |
| chr17_-_34862473 | 0.89 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr2_-_151973387 | 0.89 |
ENSMUST00000109863.1
|
Fam110a
|
family with sequence similarity 110, member A |
| chr12_+_52699297 | 0.88 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr2_-_151973840 | 0.88 |
ENSMUST00000109865.1
ENSMUST00000109864.1 |
Fam110a
|
family with sequence similarity 110, member A |
| chr6_-_131313827 | 0.88 |
ENSMUST00000049150.1
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr10_-_7780866 | 0.87 |
ENSMUST00000124838.1
ENSMUST00000039763.7 |
Ginm1
|
glycoprotein integral membrane 1 |
| chr7_-_127014585 | 0.86 |
ENSMUST00000165096.2
|
Mvp
|
major vault protein |
| chr11_+_53519920 | 0.86 |
ENSMUST00000147912.1
|
Sept8
|
septin 8 |
| chrX_+_139684980 | 0.84 |
ENSMUST00000096313.3
|
Tbc1d8b
|
TBC1 domain family, member 8B |
| chr19_+_46305682 | 0.83 |
ENSMUST00000111881.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr16_-_11176056 | 0.82 |
ENSMUST00000142389.1
ENSMUST00000138185.1 |
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr16_-_55838827 | 0.82 |
ENSMUST00000096026.2
ENSMUST00000036273.6 ENSMUST00000114457.1 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr8_-_71676674 | 0.82 |
ENSMUST00000066837.4
|
Gm9933
|
predicted gene 9933 |
| chr7_-_127930066 | 0.82 |
ENSMUST00000032988.8
|
Prss8
|
protease, serine, 8 (prostasin) |
| chr11_+_114727384 | 0.81 |
ENSMUST00000069325.7
|
Dnaic2
|
dynein, axonemal, intermediate chain 2 |
| chr8_-_99416397 | 0.81 |
ENSMUST00000155527.1
ENSMUST00000142129.1 ENSMUST00000093249.4 ENSMUST00000142475.2 ENSMUST00000128860.1 |
Cdh8
|
cadherin 8 |
| chr4_-_40239779 | 0.81 |
ENSMUST00000037907.6
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr1_+_109983006 | 0.81 |
ENSMUST00000145188.1
|
Cdh7
|
cadherin 7, type 2 |
| chr1_-_182282218 | 0.80 |
ENSMUST00000133052.1
|
Degs1
|
degenerative spermatocyte homolog 1 (Drosophila) |
| chr16_+_93683184 | 0.80 |
ENSMUST00000039620.6
|
Cbr3
|
carbonyl reductase 3 |
| chr11_-_83578496 | 0.80 |
ENSMUST00000019266.5
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr6_-_48841098 | 0.80 |
ENSMUST00000101429.4
|
Tmem176b
|
transmembrane protein 176B |
| chr11_+_53519871 | 0.79 |
ENSMUST00000120878.2
|
Sept8
|
septin 8 |
| chr7_-_19629355 | 0.78 |
ENSMUST00000049912.8
ENSMUST00000094762.3 ENSMUST00000098754.4 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr1_-_5019342 | 0.78 |
ENSMUST00000002533.8
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr17_+_35821675 | 0.78 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr15_-_75747922 | 0.77 |
ENSMUST00000062002.4
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr5_+_24413406 | 0.77 |
ENSMUST00000049346.5
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr12_+_111442453 | 0.76 |
ENSMUST00000102745.3
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr5_+_66968559 | 0.76 |
ENSMUST00000127184.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr8_+_36457548 | 0.75 |
ENSMUST00000135373.1
ENSMUST00000147525.1 |
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr1_+_109982710 | 0.75 |
ENSMUST00000112701.1
|
Cdh7
|
cadherin 7, type 2 |
| chrX_-_102908672 | 0.75 |
ENSMUST00000119624.1
ENSMUST00000033686.1 |
Dmrtc1a
|
DMRT-like family C1a |
| chr6_-_48840988 | 0.75 |
ENSMUST00000164733.1
|
Tmem176b
|
transmembrane protein 176B |
| chr8_+_71676233 | 0.74 |
ENSMUST00000110013.3
ENSMUST00000051995.7 |
Jak3
|
Janus kinase 3 |
| chr11_-_69880971 | 0.74 |
ENSMUST00000050555.3
|
Kctd11
|
potassium channel tetramerisation domain containing 11 |
| chr16_+_23290464 | 0.74 |
ENSMUST00000115335.1
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr10_-_75797528 | 0.73 |
ENSMUST00000120177.1
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr4_+_43059028 | 0.73 |
ENSMUST00000163653.1
ENSMUST00000107952.2 ENSMUST00000107953.2 |
Unc13b
|
unc-13 homolog B (C. elegans) |
| chr17_+_47436731 | 0.73 |
ENSMUST00000150819.2
|
AI661453
|
expressed sequence AI661453 |
| chr1_+_40266578 | 0.72 |
ENSMUST00000114795.1
|
Il1r1
|
interleukin 1 receptor, type I |
| chr7_-_109616548 | 0.72 |
ENSMUST00000077909.1
ENSMUST00000084738.3 |
St5
|
suppression of tumorigenicity 5 |
| chr14_-_70429072 | 0.71 |
ENSMUST00000048129.4
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr19_+_53529100 | 0.71 |
ENSMUST00000038287.6
|
Dusp5
|
dual specificity phosphatase 5 |
| chr2_-_66634952 | 0.71 |
ENSMUST00000100064.2
ENSMUST00000100063.2 |
Scn9a
|
sodium channel, voltage-gated, type IX, alpha |
| chr3_+_89421619 | 0.70 |
ENSMUST00000094378.3
ENSMUST00000137793.1 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr11_-_58502554 | 0.69 |
ENSMUST00000170501.2
ENSMUST00000081743.2 |
Olfr331
|
olfactory receptor 331 |
| chr3_-_116423930 | 0.68 |
ENSMUST00000106491.2
|
Cdc14a
|
CDC14 cell division cycle 14A |
| chr4_-_40239700 | 0.68 |
ENSMUST00000142055.1
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr18_+_51117754 | 0.67 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr4_-_101265236 | 0.67 |
ENSMUST00000149297.1
ENSMUST00000102781.3 |
Jak1
|
Janus kinase 1 |
| chr3_-_116424007 | 0.66 |
ENSMUST00000090464.4
|
Cdc14a
|
CDC14 cell division cycle 14A |
| chr2_-_152831112 | 0.65 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr4_-_135272798 | 0.64 |
ENSMUST00000037099.8
|
Clic4
|
chloride intracellular channel 4 (mitochondrial) |
| chr10_-_12964252 | 0.64 |
ENSMUST00000163425.1
ENSMUST00000042861.5 |
Stx11
|
syntaxin 11 |
| chr18_+_37484955 | 0.64 |
ENSMUST00000053856.4
|
Pcdhb17
|
protocadherin beta 17 |
| chr13_+_25056206 | 0.64 |
ENSMUST00000069614.6
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr14_+_55591708 | 0.64 |
ENSMUST00000019443.8
|
Rnf31
|
ring finger protein 31 |
| chr19_+_55741810 | 0.63 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr17_+_45555693 | 0.63 |
ENSMUST00000024742.7
|
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr8_+_114133557 | 0.63 |
ENSMUST00000073521.5
ENSMUST00000066514.6 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr3_-_121263314 | 0.63 |
ENSMUST00000029777.7
|
Tmem56
|
transmembrane protein 56 |
| chr15_-_79834323 | 0.63 |
ENSMUST00000177316.2
ENSMUST00000175858.2 |
Nptxr
|
neuronal pentraxin receptor |
| chr5_-_137116177 | 0.63 |
ENSMUST00000054384.5
ENSMUST00000152207.1 |
Trim56
|
tripartite motif-containing 56 |
| chr9_-_44526397 | 0.62 |
ENSMUST00000062215.7
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr9_-_31131817 | 0.62 |
ENSMUST00000034478.2
|
St14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr5_+_30013141 | 0.62 |
ENSMUST00000026845.7
|
Il6
|
interleukin 6 |
| chr2_-_6213033 | 0.61 |
ENSMUST00000042658.2
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr8_+_114133635 | 0.61 |
ENSMUST00000147605.1
ENSMUST00000134593.1 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr7_+_75848338 | 0.61 |
ENSMUST00000092073.5
ENSMUST00000171155.2 |
Klhl25
|
kelch-like 25 |
| chr2_+_62664279 | 0.61 |
ENSMUST00000028257.2
|
Gca
|
grancalcin |
| chr8_-_25201349 | 0.61 |
ENSMUST00000084512.4
ENSMUST00000084030.4 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr8_+_54954728 | 0.60 |
ENSMUST00000033915.7
|
Gpm6a
|
glycoprotein m6a |
| chr6_-_48841373 | 0.60 |
ENSMUST00000166247.1
|
Tmem176b
|
transmembrane protein 176B |
| chr7_-_29281977 | 0.60 |
ENSMUST00000098604.4
ENSMUST00000108236.3 |
Spint2
|
serine protease inhibitor, Kunitz type 2 |
| chr2_-_152830266 | 0.60 |
ENSMUST00000140436.1
|
Bcl2l1
|
BCL2-like 1 |
| chr14_+_65970610 | 0.60 |
ENSMUST00000127387.1
|
Clu
|
clusterin |
| chr3_-_121263159 | 0.59 |
ENSMUST00000128909.1
|
Tmem56
|
transmembrane protein 56 |
| chr6_-_129275360 | 0.59 |
ENSMUST00000032259.3
|
Cd69
|
CD69 antigen |
| chr10_+_75935573 | 0.59 |
ENSMUST00000058906.6
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr7_-_57387172 | 0.58 |
ENSMUST00000068911.6
|
Gabrg3
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 3 |
| chr1_-_172590463 | 0.58 |
ENSMUST00000065679.6
|
Slamf8
|
SLAM family member 8 |
| chr5_+_43818893 | 0.58 |
ENSMUST00000101237.4
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr10_+_69219357 | 0.58 |
ENSMUST00000172261.1
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr5_+_114568016 | 0.58 |
ENSMUST00000043650.7
|
Fam222a
|
family with sequence similarity 222, member A |
| chr3_-_113258837 | 0.58 |
ENSMUST00000098673.3
|
Amy2a5
|
amylase 2a5 |
| chr18_+_36560581 | 0.57 |
ENSMUST00000155329.2
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr2_-_156839790 | 0.57 |
ENSMUST00000134838.1
ENSMUST00000137463.1 ENSMUST00000149275.2 |
Gm14230
|
predicted gene 14230 |
| chr11_-_23770953 | 0.56 |
ENSMUST00000102864.3
|
Rel
|
reticuloendotheliosis oncogene |
| chr7_-_30861470 | 0.56 |
ENSMUST00000052700.3
|
Ffar1
|
free fatty acid receptor 1 |
| chr13_+_51846673 | 0.56 |
ENSMUST00000021903.2
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr5_-_30960236 | 0.55 |
ENSMUST00000088063.2
|
Preb
|
prolactin regulatory element binding |
| chr5_-_17835857 | 0.55 |
ENSMUST00000082367.6
|
Cd36
|
CD36 antigen |
| chr12_+_111442653 | 0.55 |
ENSMUST00000109792.1
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr1_+_58795371 | 0.55 |
ENSMUST00000027189.8
|
Casp8
|
caspase 8 |
| chr8_-_45410539 | 0.54 |
ENSMUST00000034056.4
ENSMUST00000167106.1 |
Tlr3
|
toll-like receptor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rela_Rel_Nfkb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 1.1 | 4.5 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 1.0 | 2.9 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.9 | 2.7 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.9 | 3.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.8 | 2.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.8 | 3.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.7 | 3.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.7 | 2.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.7 | 2.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.6 | 4.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.6 | 3.1 | GO:0060697 | glucosylceramide catabolic process(GO:0006680) positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.6 | 1.8 | GO:0070340 | cell surface pattern recognition receptor signaling pathway(GO:0002752) positive regulation of interleukin-18 production(GO:0032741) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.6 | 2.9 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.5 | 1.6 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.5 | 7.8 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.5 | 1.4 | GO:0009804 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.4 | 1.8 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.4 | 1.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.4 | 1.3 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.4 | 2.5 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.4 | 1.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.4 | 1.2 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.4 | 1.6 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.4 | 1.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.3 | 1.0 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.3 | 1.6 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.3 | 1.0 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 1.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.3 | 1.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 1.5 | GO:0035546 | detection of virus(GO:0009597) interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.3 | 0.6 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 0.5 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.3 | 0.8 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.3 | 0.8 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.2 | 1.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 2.0 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.7 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.7 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.2 | 1.6 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.2 | 1.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 5.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 1.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.0 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 3.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 1.0 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.2 | 0.7 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 0.5 | GO:0071726 | response to linoleic acid(GO:0070543) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 | 0.7 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.2 | 0.7 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.2 | 0.9 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 1.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.5 | GO:0015881 | creatine transport(GO:0015881) |
| 0.2 | 0.7 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.2 | 1.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 1.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 0.5 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 2.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.7 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 2.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.5 | GO:1901421 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.3 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.6 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 1.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.4 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 1.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 2.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 0.2 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.1 | 0.3 | GO:0048295 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of isotype switching to IgE isotypes(GO:0048295) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.1 | 0.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 1.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.3 | GO:0009182 | purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.1 | 0.3 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.6 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.3 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 1.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.1 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.4 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.1 | 1.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.5 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.1 | 0.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.3 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.8 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.5 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.6 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.2 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.1 | 0.2 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 1.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.6 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.3 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.5 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 1.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.7 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.2 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.1 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.6 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.1 | GO:0002925 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0039526 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) |
| 0.0 | 0.9 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.7 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin A metabolic process(GO:1901738) |
| 0.0 | 0.3 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.5 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.4 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 3.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.0 | 0.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.3 | GO:1902031 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 1.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) fatty-acyl-CoA catabolic process(GO:0036115) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.8 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.3 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 1.5 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.2 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 4.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.3 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 1.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.1 | GO:0021830 | substrate-independent telencephalic tangential migration(GO:0021826) interneuron migration from the subpallium to the cortex(GO:0021830) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.9 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 3.7 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.5 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:2000251 | regulation of establishment of cell polarity(GO:2000114) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.6 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.2 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.5 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.8 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 2.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.4 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.4 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.6 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.6 | GO:0010524 | positive regulation of calcium ion transport into cytosol(GO:0010524) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0060853 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) regulation of cell adhesion involved in heart morphogenesis(GO:0061344) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) |
| 0.0 | 0.2 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.9 | 2.7 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.3 | 1.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 3.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 3.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 2.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 2.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 3.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.2 | 1.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 0.4 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 0.5 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 0.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 0.5 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.2 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 2.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.7 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 4.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 15.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 12.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 3.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 3.6 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 2.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.9 | 2.7 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.8 | 10.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.8 | 3.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.5 | 1.9 | GO:0042903 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.4 | 3.7 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.4 | 1.8 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.3 | 1.8 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.3 | 2.0 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.3 | 2.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 2.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 1.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 0.8 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 0.7 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 0.7 | GO:0016824 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.2 | 0.7 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 1.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 3.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 0.8 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.2 | 1.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 0.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.5 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.2 | 7.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 0.6 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 0.5 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.2 | 0.8 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 1.2 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.1 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 3.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.9 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 1.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 1.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.4 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 2.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0051998 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.2 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 5.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.0 | GO:0047555 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 4.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0051378 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 1.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 2.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 3.0 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 6.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 6.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 8.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 10.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 3.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 2.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 4.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 3.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 5.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 4.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 3.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.3 | 3.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 5.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 1.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 10.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 9.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 1.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 1.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 2.8 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.1 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 5.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.2 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 3.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |