Project
avrg: 2D miR_HR1_12
Navigation
Downloads
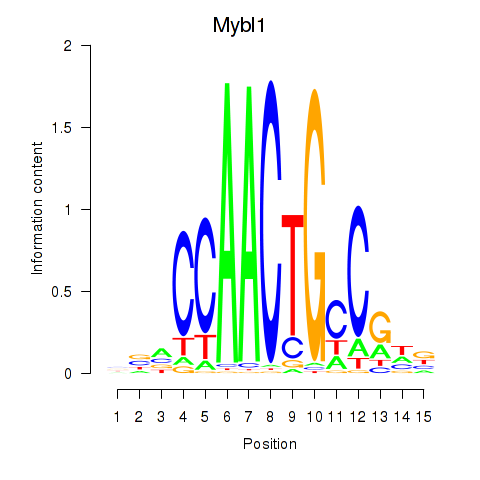
Results for Mybl1
Z-value: 5.09
Transcription factors associated with Mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl1
|
ENSMUSG00000025912.10 | myeloblastosis oncogene-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl1 | mm10_v2_chr1_-_9700209_9700329 | 0.96 | 4.1e-07 | Click! |
Activity profile of Mybl1 motif
Sorted Z-values of Mybl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_118814195 | 9.50 |
ENSMUST00000110842.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr2_+_118814237 | 8.54 |
ENSMUST00000028803.7
ENSMUST00000126045.1 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr14_+_65806066 | 8.21 |
ENSMUST00000139644.1
|
Pbk
|
PDZ binding kinase |
| chr2_+_118813995 | 8.03 |
ENSMUST00000134661.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr4_-_118437331 | 7.55 |
ENSMUST00000006565.6
|
Cdc20
|
cell division cycle 20 |
| chrX_-_102157065 | 6.82 |
ENSMUST00000056904.2
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr14_+_46760526 | 5.70 |
ENSMUST00000067426.4
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr7_+_79660196 | 5.65 |
ENSMUST00000035977.7
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr6_+_124829540 | 5.56 |
ENSMUST00000150120.1
|
Cdca3
|
cell division cycle associated 3 |
| chr4_-_117182623 | 5.47 |
ENSMUST00000065896.2
|
Kif2c
|
kinesin family member 2C |
| chr1_+_57995971 | 5.35 |
ENSMUST00000027202.8
|
Sgol2
|
shugoshin-like 2 (S. pombe) |
| chr5_+_123749696 | 5.25 |
ENSMUST00000031366.7
|
Kntc1
|
kinetochore associated 1 |
| chr6_+_124829582 | 5.09 |
ENSMUST00000024270.7
|
Cdca3
|
cell division cycle associated 3 |
| chr7_+_126862431 | 4.80 |
ENSMUST00000132808.1
|
Hirip3
|
HIRA interacting protein 3 |
| chr19_-_10101501 | 4.40 |
ENSMUST00000025567.7
|
Fads2
|
fatty acid desaturase 2 |
| chr13_-_49652714 | 4.39 |
ENSMUST00000021818.7
|
Cenpp
|
centromere protein P |
| chr1_+_9547948 | 4.35 |
ENSMUST00000144177.1
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr4_-_43499608 | 4.24 |
ENSMUST00000136005.1
ENSMUST00000054538.6 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr1_-_169531447 | 4.15 |
ENSMUST00000111368.1
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr15_-_83367267 | 4.04 |
ENSMUST00000100370.1
ENSMUST00000178628.1 |
1700001L05Rik
|
RIKEN cDNA 1700001L05 gene |
| chr14_+_65805832 | 3.96 |
ENSMUST00000022612.3
|
Pbk
|
PDZ binding kinase |
| chr5_+_110286306 | 3.86 |
ENSMUST00000007296.5
ENSMUST00000112482.1 |
Pole
|
polymerase (DNA directed), epsilon |
| chr16_+_93883895 | 3.74 |
ENSMUST00000023666.4
ENSMUST00000117099.1 ENSMUST00000142316.1 |
Chaf1b
|
chromatin assembly factor 1, subunit B (p60) |
| chr14_+_115042752 | 3.68 |
ENSMUST00000134140.2
|
Mir17hg
|
Mir17 host gene 1 (non-protein coding) |
| chr7_-_62420139 | 3.68 |
ENSMUST00000094340.3
|
Mkrn3
|
makorin, ring finger protein, 3 |
| chr1_-_44101982 | 3.66 |
ENSMUST00000127923.1
|
Tex30
|
testis expressed 30 |
| chr3_-_69044697 | 3.58 |
ENSMUST00000136512.1
ENSMUST00000143454.1 ENSMUST00000107802.1 |
Trim59
|
tripartite motif-containing 59 |
| chr17_-_26095487 | 3.55 |
ENSMUST00000025007.5
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr15_-_83367006 | 3.55 |
ENSMUST00000179705.1
|
1700001L05Rik
|
RIKEN cDNA 1700001L05 gene |
| chr1_+_153425162 | 3.43 |
ENSMUST00000042373.5
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr1_-_191575534 | 3.39 |
ENSMUST00000027933.5
|
Dtl
|
denticleless homolog (Drosophila) |
| chr12_+_69168808 | 3.32 |
ENSMUST00000110621.1
|
Lrr1
|
leucine rich repeat protein 1 |
| chr8_+_83715504 | 3.26 |
ENSMUST00000109810.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr19_+_5024006 | 3.25 |
ENSMUST00000025826.5
|
Slc29a2
|
solute carrier family 29 (nucleoside transporters), member 2 |
| chr6_-_113600645 | 3.19 |
ENSMUST00000035870.4
|
Fancd2os
|
Fancd2 opposite strand |
| chr8_+_83955507 | 3.02 |
ENSMUST00000005607.8
|
Asf1b
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae) |
| chrX_-_74353575 | 2.99 |
ENSMUST00000114152.1
ENSMUST00000114153.1 ENSMUST00000015433.3 |
Lage3
|
L antigen family, member 3 |
| chr10_-_80855187 | 2.90 |
ENSMUST00000035775.8
|
Lsm7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr9_-_88522876 | 2.87 |
ENSMUST00000180563.2
ENSMUST00000183030.1 ENSMUST00000182232.1 |
Snhg5
|
small nucleolar RNA host gene 5 |
| chr12_-_99883429 | 2.86 |
ENSMUST00000046485.3
|
Efcab11
|
EF-hand calcium binding domain 11 |
| chrX_+_71364901 | 2.85 |
ENSMUST00000132837.1
|
Mtmr1
|
myotubularin related protein 1 |
| chr19_-_41802028 | 2.84 |
ENSMUST00000026150.8
ENSMUST00000177495.1 ENSMUST00000163265.1 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr3_-_88950271 | 2.84 |
ENSMUST00000174402.1
ENSMUST00000174077.1 |
Dap3
|
death associated protein 3 |
| chr9_+_53771499 | 2.83 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr3_-_88410295 | 2.80 |
ENSMUST00000056370.7
|
Pmf1
|
polyamine-modulated factor 1 |
| chr2_+_119112793 | 2.80 |
ENSMUST00000140939.1
ENSMUST00000028795.3 |
Rad51
|
RAD51 homolog |
| chr17_+_29093763 | 2.76 |
ENSMUST00000023829.6
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr16_+_35983424 | 2.73 |
ENSMUST00000173555.1
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr3_-_108402589 | 2.67 |
ENSMUST00000147565.1
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) |
| chr5_+_98854434 | 2.61 |
ENSMUST00000031278.4
|
Bmp3
|
bone morphogenetic protein 3 |
| chr3_+_68572245 | 2.55 |
ENSMUST00000170788.2
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_-_45434590 | 2.54 |
ENSMUST00000107771.3
ENSMUST00000141761.1 |
Ruvbl2
|
RuvB-like protein 2 |
| chr2_+_121506715 | 2.53 |
ENSMUST00000028676.5
|
Wdr76
|
WD repeat domain 76 |
| chr2_+_109280738 | 2.52 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr9_+_95857597 | 2.50 |
ENSMUST00000034980.7
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr8_+_83715177 | 2.49 |
ENSMUST00000019576.8
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr3_-_88949906 | 2.48 |
ENSMUST00000172942.1
ENSMUST00000107491.4 |
Dap3
|
death associated protein 3 |
| chr18_+_34624621 | 2.47 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr1_-_37719782 | 2.44 |
ENSMUST00000160589.1
|
2010300C02Rik
|
RIKEN cDNA 2010300C02 gene |
| chr11_+_29130733 | 2.40 |
ENSMUST00000020756.8
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr3_+_10012548 | 2.39 |
ENSMUST00000029046.8
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr13_-_38658991 | 2.36 |
ENSMUST00000001757.7
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr13_-_54590047 | 2.34 |
ENSMUST00000148222.1
ENSMUST00000026987.5 |
Nop16
|
NOP16 nucleolar protein |
| chr17_-_84790517 | 2.30 |
ENSMUST00000112308.2
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr12_-_79192248 | 2.29 |
ENSMUST00000161204.1
|
Rdh11
|
retinol dehydrogenase 11 |
| chr1_-_44102414 | 2.29 |
ENSMUST00000143327.1
ENSMUST00000133677.1 |
Tex30
|
testis expressed 30 |
| chr7_-_141279121 | 2.27 |
ENSMUST00000167790.1
ENSMUST00000046156.6 |
Sct
|
secretin |
| chr5_-_110286159 | 2.26 |
ENSMUST00000031472.5
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr10_-_63244135 | 2.26 |
ENSMUST00000054837.3
|
1700120B22Rik
|
RIKEN cDNA 1700120B22 gene |
| chr6_-_72439549 | 2.25 |
ENSMUST00000059472.8
|
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr13_-_24761440 | 2.25 |
ENSMUST00000176890.1
ENSMUST00000175689.1 |
Gmnn
|
geminin |
| chr18_-_73815392 | 2.22 |
ENSMUST00000025439.3
|
Me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr1_-_33669745 | 2.21 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr2_+_150909565 | 2.20 |
ENSMUST00000028948.4
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr6_+_79818031 | 2.17 |
ENSMUST00000179797.1
|
Gm20594
|
predicted gene, 20594 |
| chr4_+_107367757 | 2.16 |
ENSMUST00000139560.1
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr5_+_38220470 | 2.15 |
ENSMUST00000087514.2
ENSMUST00000130721.1 ENSMUST00000123207.1 ENSMUST00000132190.1 ENSMUST00000152066.1 ENSMUST00000155300.1 |
Lyar
|
Ly1 antibody reactive clone |
| chr13_+_108316395 | 2.15 |
ENSMUST00000171178.1
|
Depdc1b
|
DEP domain containing 1B |
| chr11_-_61453992 | 2.13 |
ENSMUST00000060255.7
ENSMUST00000054927.7 ENSMUST00000102661.3 |
Rnf112
|
ring finger protein 112 |
| chr3_+_145576196 | 2.11 |
ENSMUST00000098534.4
|
Znhit6
|
zinc finger, HIT type 6 |
| chr7_-_19399859 | 2.09 |
ENSMUST00000047170.3
ENSMUST00000108459.2 |
Klc3
|
kinesin light chain 3 |
| chr6_+_136808248 | 2.08 |
ENSMUST00000074556.4
|
H2afj
|
H2A histone family, member J |
| chr1_-_60098104 | 2.06 |
ENSMUST00000143342.1
|
Wdr12
|
WD repeat domain 12 |
| chr16_+_20629799 | 2.05 |
ENSMUST00000003898.5
|
Ece2
|
endothelin converting enzyme 2 |
| chr1_-_93801840 | 2.04 |
ENSMUST00000112890.2
ENSMUST00000027503.7 |
Dtymk
|
deoxythymidylate kinase |
| chr1_+_9908638 | 2.02 |
ENSMUST00000171802.1
ENSMUST00000052843.5 ENSMUST00000125294.2 ENSMUST00000140948.2 |
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr2_+_30807826 | 2.02 |
ENSMUST00000041830.3
ENSMUST00000152374.1 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr16_+_20629842 | 2.01 |
ENSMUST00000122306.1
ENSMUST00000133344.1 |
Ece2
|
endothelin converting enzyme 2 |
| chr6_+_35177610 | 1.98 |
ENSMUST00000170234.1
|
Nup205
|
nucleoporin 205 |
| chr8_-_48555846 | 1.98 |
ENSMUST00000110345.1
ENSMUST00000110343.1 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr17_-_27635238 | 1.98 |
ENSMUST00000025052.6
ENSMUST00000114882.1 |
Rps10
|
ribosomal protein S10 |
| chr13_-_24761861 | 1.97 |
ENSMUST00000006898.3
ENSMUST00000110382.2 |
Gmnn
|
geminin |
| chr1_-_167285110 | 1.96 |
ENSMUST00000027839.8
|
Uck2
|
uridine-cytidine kinase 2 |
| chr4_-_132533488 | 1.96 |
ENSMUST00000152993.1
ENSMUST00000067496.6 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr2_+_163054682 | 1.96 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr11_-_69921190 | 1.94 |
ENSMUST00000108607.1
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_-_44102362 | 1.93 |
ENSMUST00000147571.1
ENSMUST00000027215.5 ENSMUST00000147661.1 |
Tex30
|
testis expressed 30 |
| chr5_+_38220628 | 1.92 |
ENSMUST00000114106.1
|
Lyar
|
Ly1 antibody reactive clone |
| chr1_-_21961581 | 1.91 |
ENSMUST00000029667.6
ENSMUST00000173058.1 ENSMUST00000173404.1 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr3_+_88043098 | 1.89 |
ENSMUST00000166021.1
ENSMUST00000029707.7 |
Gpatch4
|
G patch domain containing 4 |
| chr1_-_44102433 | 1.89 |
ENSMUST00000129702.1
ENSMUST00000149502.1 ENSMUST00000156392.1 ENSMUST00000150911.1 |
Tex30
|
testis expressed 30 |
| chr7_+_132610620 | 1.89 |
ENSMUST00000033241.5
|
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr11_+_23256566 | 1.87 |
ENSMUST00000136235.1
|
Xpo1
|
exportin 1, CRM1 homolog (yeast) |
| chr4_+_104913456 | 1.86 |
ENSMUST00000106803.2
ENSMUST00000106804.1 |
1700024P16Rik
|
RIKEN cDNA 1700024P16 gene |
| chr1_-_60098135 | 1.86 |
ENSMUST00000141417.1
ENSMUST00000122038.1 |
Wdr12
|
WD repeat domain 12 |
| chr1_+_55237177 | 1.85 |
ENSMUST00000061334.8
|
Mars2
|
methionine-tRNA synthetase 2 (mitochondrial) |
| chr14_-_49245389 | 1.85 |
ENSMUST00000130853.1
ENSMUST00000022398.7 |
1700011H14Rik
|
RIKEN cDNA 1700011H14 gene |
| chr11_-_99155067 | 1.81 |
ENSMUST00000103134.3
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr8_+_22411340 | 1.81 |
ENSMUST00000033934.3
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr12_+_5411641 | 1.81 |
ENSMUST00000163627.1
|
2810032G03Rik
|
RIKEN cDNA 2810032G03 gene |
| chr15_+_78406695 | 1.80 |
ENSMUST00000167140.1
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr12_-_91746020 | 1.78 |
ENSMUST00000166967.1
|
Ston2
|
stonin 2 |
| chrX_+_48108912 | 1.78 |
ENSMUST00000114998.1
ENSMUST00000115000.3 |
Xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr15_+_78406777 | 1.78 |
ENSMUST00000169133.1
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr4_-_44167988 | 1.77 |
ENSMUST00000143337.1
|
Rnf38
|
ring finger protein 38 |
| chr10_-_128565827 | 1.77 |
ENSMUST00000131728.1
ENSMUST00000026425.6 |
Pa2g4
|
proliferation-associated 2G4 |
| chr17_+_28328471 | 1.77 |
ENSMUST00000042334.8
|
Rpl10a
|
ribosomal protein L10A |
| chrX_+_73673150 | 1.75 |
ENSMUST00000033752.7
ENSMUST00000114467.2 |
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr4_+_128993224 | 1.75 |
ENSMUST00000030583.6
ENSMUST00000102604.4 |
Ak2
|
adenylate kinase 2 |
| chr1_-_44102341 | 1.75 |
ENSMUST00000128190.1
|
Tex30
|
testis expressed 30 |
| chr7_-_4778141 | 1.74 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr1_-_75219245 | 1.73 |
ENSMUST00000079464.6
|
Tuba4a
|
tubulin, alpha 4A |
| chr8_-_120634379 | 1.72 |
ENSMUST00000123927.1
|
1190005I06Rik
|
RIKEN cDNA 1190005I06 gene |
| chr9_-_106447584 | 1.71 |
ENSMUST00000171678.1
ENSMUST00000048685.6 ENSMUST00000171925.1 |
Abhd14a
|
abhydrolase domain containing 14A |
| chrX_-_97377190 | 1.71 |
ENSMUST00000037353.3
|
Eda2r
|
ectodysplasin A2 receptor |
| chr5_-_130003000 | 1.70 |
ENSMUST00000026613.7
|
Gusb
|
glucuronidase, beta |
| chr7_-_127260677 | 1.69 |
ENSMUST00000035276.4
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr11_+_119942763 | 1.67 |
ENSMUST00000026436.3
ENSMUST00000106231.1 ENSMUST00000075180.5 ENSMUST00000103021.3 ENSMUST00000106233.1 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr2_+_130274424 | 1.65 |
ENSMUST00000103198.4
|
Nop56
|
NOP56 ribonucleoprotein |
| chr3_+_87906842 | 1.64 |
ENSMUST00000159492.1
|
Hdgf
|
hepatoma-derived growth factor |
| chr11_-_69921329 | 1.64 |
ENSMUST00000108613.3
ENSMUST00000043419.3 ENSMUST00000070996.4 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_180568913 | 1.63 |
ENSMUST00000027777.6
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
| chr17_+_87672523 | 1.63 |
ENSMUST00000172855.1
|
Msh2
|
mutS homolog 2 (E. coli) |
| chr2_+_119034783 | 1.62 |
ENSMUST00000028796.1
|
Rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr19_-_4839286 | 1.61 |
ENSMUST00000037246.5
|
Ccs
|
copper chaperone for superoxide dismutase |
| chr13_-_54766553 | 1.58 |
ENSMUST00000036825.7
|
Sncb
|
synuclein, beta |
| chr4_+_108847827 | 1.58 |
ENSMUST00000102738.2
|
Kti12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr17_-_35175995 | 1.56 |
ENSMUST00000173324.1
|
Aif1
|
allograft inflammatory factor 1 |
| chr13_+_49682100 | 1.54 |
ENSMUST00000165316.1
ENSMUST00000047363.7 |
Iars
|
isoleucine-tRNA synthetase |
| chr8_+_83715239 | 1.53 |
ENSMUST00000172396.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr8_-_105851981 | 1.53 |
ENSMUST00000040776.4
|
Cenpt
|
centromere protein T |
| chr19_-_40271506 | 1.53 |
ENSMUST00000068439.6
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr11_+_101325063 | 1.52 |
ENSMUST00000041095.7
ENSMUST00000107264.1 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr17_+_88668660 | 1.51 |
ENSMUST00000024970.4
ENSMUST00000161481.1 |
Gtf2a1l
|
general transcription factor IIA, 1-like |
| chr2_+_151543877 | 1.51 |
ENSMUST00000142271.1
|
Fkbp1a
|
FK506 binding protein 1a |
| chr2_-_105399286 | 1.50 |
ENSMUST00000006128.6
|
Rcn1
|
reticulocalbin 1 |
| chr18_+_34736359 | 1.45 |
ENSMUST00000105038.2
|
Gm3550
|
predicted gene 3550 |
| chr11_+_68901538 | 1.45 |
ENSMUST00000073471.6
ENSMUST00000101014.2 ENSMUST00000128952.1 ENSMUST00000167436.1 |
Rpl26
|
ribosomal protein L26 |
| chr2_+_121506748 | 1.45 |
ENSMUST00000099473.3
ENSMUST00000110602.2 |
Wdr76
|
WD repeat domain 76 |
| chr10_+_127677064 | 1.45 |
ENSMUST00000118612.1
ENSMUST00000048099.4 |
Tmem194
|
transmembrane protein 194 |
| chr4_+_155891822 | 1.44 |
ENSMUST00000105584.3
ENSMUST00000079031.5 |
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr9_+_106281061 | 1.44 |
ENSMUST00000072206.6
|
Poc1a
|
POC1 centriolar protein homolog A (Chlamydomonas) |
| chr2_+_84826997 | 1.43 |
ENSMUST00000028470.3
|
Timm10
|
translocase of inner mitochondrial membrane 10 |
| chr13_-_23762378 | 1.43 |
ENSMUST00000091701.2
|
Hist1h3a
|
histone cluster 1, H3a |
| chr7_-_57509995 | 1.42 |
ENSMUST00000068456.6
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr18_+_67800101 | 1.41 |
ENSMUST00000025425.5
|
Cep192
|
centrosomal protein 192 |
| chr1_+_171018920 | 1.41 |
ENSMUST00000078825.4
|
Fcgr4
|
Fc receptor, IgG, low affinity IV |
| chr2_+_19371636 | 1.40 |
ENSMUST00000023856.8
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr7_-_118116171 | 1.40 |
ENSMUST00000131374.1
|
Rps15a
|
ribosomal protein S15A |
| chr11_+_68692097 | 1.40 |
ENSMUST00000018887.8
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr14_+_24490678 | 1.39 |
ENSMUST00000169826.1
ENSMUST00000112384.3 |
Rps24
|
ribosomal protein S24 |
| chr8_-_68121527 | 1.38 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chr11_+_68691906 | 1.38 |
ENSMUST00000102611.3
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr7_-_118116128 | 1.36 |
ENSMUST00000128482.1
ENSMUST00000131840.1 |
Rps15a
|
ribosomal protein S15A |
| chr14_-_13961202 | 1.36 |
ENSMUST00000065865.8
|
Thoc7
|
THO complex 7 homolog (Drosophila) |
| chr8_+_69226343 | 1.35 |
ENSMUST00000110216.1
|
Zfp930
|
zinc finger protein 930 |
| chr13_-_110280103 | 1.35 |
ENSMUST00000167824.1
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chrX_-_97377150 | 1.34 |
ENSMUST00000113832.1
|
Eda2r
|
ectodysplasin A2 receptor |
| chr6_+_89643982 | 1.34 |
ENSMUST00000000828.6
ENSMUST00000101171.1 |
Txnrd3
|
thioredoxin reductase 3 |
| chr8_-_13975032 | 1.33 |
ENSMUST00000145695.1
|
Tdrp
|
testis development related protein |
| chr3_+_116594959 | 1.33 |
ENSMUST00000029571.8
|
Sass6
|
spindle assembly 6 homolog (C. elegans) |
| chr1_-_179546261 | 1.32 |
ENSMUST00000027769.5
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr5_+_146384947 | 1.32 |
ENSMUST00000110600.1
ENSMUST00000016143.7 |
Wasf3
|
WAS protein family, member 3 |
| chr1_-_55088156 | 1.32 |
ENSMUST00000127861.1
ENSMUST00000144077.1 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr13_-_77131276 | 1.32 |
ENSMUST00000159300.1
|
Ankrd32
|
ankyrin repeat domain 32 |
| chr2_-_84670659 | 1.32 |
ENSMUST00000102646.1
ENSMUST00000102647.3 |
2700094K13Rik
|
RIKEN cDNA 2700094K13 gene |
| chr7_+_65644884 | 1.31 |
ENSMUST00000032728.8
|
Tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr4_-_137766474 | 1.30 |
ENSMUST00000139951.1
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr2_-_5895319 | 1.30 |
ENSMUST00000026926.4
ENSMUST00000102981.3 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr15_-_80264276 | 1.29 |
ENSMUST00000052499.7
|
Rps19bp1
|
ribosomal protein S19 binding protein 1 |
| chr14_+_63860290 | 1.29 |
ENSMUST00000022528.4
|
Pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr7_+_43437073 | 1.29 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr5_-_48754521 | 1.29 |
ENSMUST00000101214.2
ENSMUST00000176191.1 |
Kcnip4
|
Kv channel interacting protein 4 |
| chr2_-_26380600 | 1.29 |
ENSMUST00000114115.2
ENSMUST00000035427.4 |
Snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr16_-_36071515 | 1.28 |
ENSMUST00000004057.7
|
Fam162a
|
family with sequence similarity 162, member A |
| chr11_-_106998483 | 1.26 |
ENSMUST00000124541.1
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr8_-_71272367 | 1.25 |
ENSMUST00000110071.2
|
Haus8
|
4HAUS augmin-like complex, subunit 8 |
| chr12_+_3426857 | 1.25 |
ENSMUST00000111215.3
ENSMUST00000092003.5 ENSMUST00000144247.2 ENSMUST00000153102.2 |
Asxl2
|
additional sex combs like 2 (Drosophila) |
| chr10_+_76147451 | 1.25 |
ENSMUST00000020450.3
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr2_-_103796989 | 1.24 |
ENSMUST00000111147.1
|
Caprin1
|
cell cycle associated protein 1 |
| chr11_-_33513626 | 1.23 |
ENSMUST00000037522.7
|
Ranbp17
|
RAN binding protein 17 |
| chr7_-_4812351 | 1.22 |
ENSMUST00000079496.7
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chrX_+_56894372 | 1.22 |
ENSMUST00000136396.1
|
Gpr112
|
G protein-coupled receptor 112 |
| chr2_+_31950257 | 1.22 |
ENSMUST00000001920.7
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr1_+_178187721 | 1.21 |
ENSMUST00000159284.1
|
Desi2
|
desumoylating isopeptidase 2 |
| chr12_+_65036319 | 1.20 |
ENSMUST00000120580.1
|
Prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr11_+_68692070 | 1.20 |
ENSMUST00000108673.1
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr5_-_134678190 | 1.20 |
ENSMUST00000111233.1
|
Limk1
|
LIM-domain containing, protein kinase |
| chr15_+_52712434 | 1.20 |
ENSMUST00000037115.7
|
Med30
|
mediator complex subunit 30 |
| chr7_+_126861947 | 1.20 |
ENSMUST00000037248.3
|
Hirip3
|
HIRA interacting protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 1.4 | 26.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.3 | 4.0 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.3 | 3.9 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 1.2 | 4.7 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 1.1 | 5.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 5.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.8 | 2.5 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.8 | 2.4 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.8 | 2.3 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.8 | 3.0 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.7 | 4.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.7 | 2.1 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.7 | 5.7 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.7 | 2.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.7 | 3.4 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.7 | 2.0 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.6 | 2.5 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.6 | 1.8 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.6 | 3.6 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.6 | 1.8 | GO:0015881 | creatine transport(GO:0015881) |
| 0.6 | 2.3 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.6 | 2.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.6 | 2.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 1.6 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.5 | 1.5 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.5 | 2.0 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.5 | 1.4 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.4 | 1.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.4 | 1.3 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.4 | 1.7 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.4 | 1.3 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.4 | 1.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 1.6 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) gene conversion(GO:0035822) |
| 0.4 | 2.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.4 | 1.6 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 | 1.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.4 | 2.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.4 | 0.7 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.4 | 1.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.4 | 2.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.4 | 1.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 2.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.3 | 2.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.3 | 0.7 | GO:0090343 | regulation of fermentation(GO:0043465) positive regulation of cell aging(GO:0090343) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.3 | 2.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 3.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 1.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 2.2 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 1.8 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.3 | 3.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 1.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 1.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 2.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 1.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 4.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.3 | 1.3 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.3 | 3.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.3 | 1.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.3 | 1.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 | 1.8 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.3 | 0.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 5.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 2.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.2 | 2.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 1.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 1.2 | GO:0032202 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 0.7 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.2 | 1.6 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 | 0.7 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.2 | 0.9 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.2 | 1.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 7.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.7 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 | 0.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 0.9 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 0.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 0.6 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 0.6 | GO:0060599 | hypophysis morphogenesis(GO:0048850) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.2 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.6 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.2 | 1.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.2 | 0.6 | GO:0031064 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) negative regulation of histone deacetylation(GO:0031064) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.2 | 0.6 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.2 | 1.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.8 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.2 | 3.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 14.4 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.2 | 1.0 | GO:0060244 | amygdala development(GO:0021764) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 1.5 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 1.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 0.4 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.2 | 7.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.2 | 0.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.2 | 0.7 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 1.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 3.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.9 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 | 0.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.2 | 1.5 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.2 | 0.8 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.2 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.2 | 1.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 1.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 1.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 2.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 2.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.6 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 1.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 3.3 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 1.1 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 3.4 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 2.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 2.5 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 1.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.4 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 1.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 3.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.5 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 0.6 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 1.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 2.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 1.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.4 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 1.2 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.1 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 4.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.5 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.4 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.3 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 0.2 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.7 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 4.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.2 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.1 | 0.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.6 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 0.7 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 1.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) CD8-positive, alpha-beta T cell extravasation(GO:0035697) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 2.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.7 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.9 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.1 | 1.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0060744 | retinal pigment epithelium development(GO:0003406) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 1.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.2 | GO:0021941 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.1 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.7 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.1 | GO:0043278 | response to morphine(GO:0043278) |
| 0.1 | 0.2 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 1.4 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 8.0 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 0.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.1 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.1 | 0.7 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 0.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 2.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 1.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.6 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 2.0 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.3 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.1 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 1.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 2.3 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.7 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.2 | GO:2000758 | positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 4.7 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.1 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.1 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 1.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.5 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.6 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 7.0 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 19.6 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.7 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 3.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) cristae formation(GO:0042407) |
| 0.0 | 1.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 3.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0009814 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.3 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.1 | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 1.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.0 | 0.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 3.4 | GO:2001252 | positive regulation of chromosome organization(GO:2001252) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 1.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 1.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.8 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:2000757 | negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.3 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 2.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0031554 | termination of RNA polymerase II transcription(GO:0006369) regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.0 | 0.7 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.4 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 2.5 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.9 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.9 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.2 | 3.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.9 | 2.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.9 | 31.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.8 | 3.9 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.8 | 2.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.7 | 2.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.7 | 8.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.7 | 4.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.7 | 4.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.5 | 3.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 4.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.5 | 2.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.5 | 3.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.5 | 2.8 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.4 | 2.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 2.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 1.2 | GO:0000799 | nuclear condensin complex(GO:0000799) germinal vesicle(GO:0042585) |
| 0.4 | 2.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 3.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 2.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 9.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 1.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.3 | 1.3 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.3 | 2.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 1.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 0.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.3 | 1.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.8 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 3.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 8.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 0.7 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.2 | 1.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.9 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 2.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 0.8 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.2 | 1.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 1.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 2.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 1.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 2.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 0.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 3.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.9 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 11.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.4 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 6.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 1.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 2.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.7 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 2.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 9.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 2.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 9.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 3.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 2.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 2.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 1.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 7.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.9 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.1 | 5.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.9 | 2.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.7 | 2.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.7 | 4.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.7 | 3.6 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.7 | 4.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.7 | 2.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.6 | 1.8 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.6 | 1.8 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.6 | 4.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.5 | 1.5 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.5 | 1.5 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.5 | 1.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 8.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 3.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 1.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.4 | 3.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 1.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 1.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 1.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.4 | 2.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 2.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.4 | 2.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 5.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.4 | 2.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 2.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 1.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 3.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 2.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 2.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 3.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 1.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 3.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 1.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 2.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 0.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.2 | 1.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 0.7 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 6.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 1.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 2.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 1.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 4.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 1.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.8 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 1.0 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.2 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.6 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 3.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 1.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.6 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 0.6 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 1.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.4 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.7 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 2.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.9 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.1 | 1.7 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 0.9 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.6 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 5.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 3.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.9 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 17.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 3.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 5.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.7 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.2 | GO:0032029 | myosin tail binding(GO:0032029) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 2.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 7.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.9 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.4 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.5 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 5.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 2.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 5.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 2.0 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 3.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 17.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 7.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 7.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 3.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 4.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 3.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 47.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.4 | 2.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 4.9 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 7.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 7.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 3.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 2.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 6.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 4.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 5.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 3.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.2 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.1 | 2.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 6.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.9 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 4.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 4.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 4.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 7.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 3.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 3.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |