Project
2D miR_HR1_12
Navigation
Downloads
Results for Stat5a
Z-value: 1.97
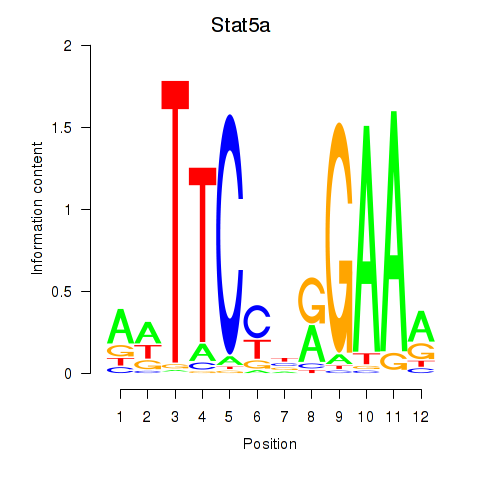
Transcription factors associated with Stat5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat5a
|
ENSMUSG00000004043.8 | signal transducer and activator of transcription 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat5a | mm10_v2_chr11_+_100860326_100860345 | -0.26 | 4.1e-01 | Click! |
Activity profile of Stat5a motif
Sorted Z-values of Stat5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_78468863 | 5.27 |
ENSMUST00000032089.2
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr6_+_78370877 | 5.19 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr11_+_69964758 | 4.17 |
ENSMUST00000108597.1
ENSMUST00000060651.5 ENSMUST00000108596.1 |
Cldn7
|
claudin 7 |
| chr6_-_41377604 | 3.72 |
ENSMUST00000096003.5
|
Prss3
|
protease, serine, 3 |
| chr6_+_78380700 | 3.63 |
ENSMUST00000101272.1
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr7_+_43995833 | 3.53 |
ENSMUST00000007156.4
|
Klk1b11
|
kallikrein 1-related peptidase b11 |
| chr9_+_107296843 | 3.23 |
ENSMUST00000167072.1
|
Cish
|
cytokine inducible SH2-containing protein |
| chr5_-_108795352 | 3.08 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr10_+_87521795 | 2.84 |
ENSMUST00000020241.8
|
Pah
|
phenylalanine hydroxylase |
| chr6_+_41354105 | 2.79 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr7_-_46715676 | 2.63 |
ENSMUST00000006956.7
|
Saa3
|
serum amyloid A 3 |
| chr10_+_87521920 | 2.60 |
ENSMUST00000142088.1
|
Pah
|
phenylalanine hydroxylase |
| chr14_+_30886476 | 2.54 |
ENSMUST00000006703.6
ENSMUST00000078490.5 ENSMUST00000120269.2 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr9_+_107296682 | 2.43 |
ENSMUST00000168260.1
|
Cish
|
cytokine inducible SH2-containing protein |
| chr6_-_41446062 | 2.31 |
ENSMUST00000095999.5
|
Gm10334
|
predicted gene 10334 |
| chr7_+_44216456 | 2.29 |
ENSMUST00000074359.2
|
Klk1b5
|
kallikrein 1-related peptidase b5 |
| chr6_-_78378851 | 2.28 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr11_+_69965396 | 2.22 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr10_+_128267997 | 2.18 |
ENSMUST00000050901.2
|
Apof
|
apolipoprotein F |
| chr6_+_41521782 | 2.16 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr6_+_40964760 | 2.10 |
ENSMUST00000076638.5
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr5_+_104435112 | 2.08 |
ENSMUST00000031243.8
ENSMUST00000086833.6 ENSMUST00000112748.1 ENSMUST00000112746.1 ENSMUST00000145084.1 ENSMUST00000132457.1 |
Spp1
|
secreted phosphoprotein 1 |
| chr18_-_36726730 | 2.03 |
ENSMUST00000061829.6
|
Cd14
|
CD14 antigen |
| chr7_+_44198191 | 2.01 |
ENSMUST00000085450.2
|
Klk1b3
|
kallikrein 1-related peptidase b3 |
| chr6_-_41035501 | 1.93 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr11_-_109722214 | 1.90 |
ENSMUST00000020938.7
|
Fam20a
|
family with sequence similarity 20, member A |
| chr6_+_78425973 | 1.83 |
ENSMUST00000079926.5
|
Reg1
|
regenerating islet-derived 1 |
| chr2_+_164403194 | 1.83 |
ENSMUST00000017151.1
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr8_+_94810446 | 1.79 |
ENSMUST00000034232.1
|
Ccl17
|
chemokine (C-C motif) ligand 17 |
| chr14_+_30886521 | 1.78 |
ENSMUST00000168782.1
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr18_-_43737186 | 1.72 |
ENSMUST00000025381.2
|
Spink3
|
serine peptidase inhibitor, Kazal type 3 |
| chr3_+_19957037 | 1.67 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr11_+_69966896 | 1.67 |
ENSMUST00000151515.1
|
Cldn7
|
claudin 7 |
| chr10_+_87521954 | 1.58 |
ENSMUST00000143624.1
|
Pah
|
phenylalanine hydroxylase |
| chr9_+_107295928 | 1.57 |
ENSMUST00000085102.5
|
Cish
|
cytokine inducible SH2-containing protein |
| chr19_+_56287911 | 1.56 |
ENSMUST00000095948.4
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr7_-_126625676 | 1.52 |
ENSMUST00000032961.3
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr4_-_149454971 | 1.52 |
ENSMUST00000030848.2
|
Rbp7
|
retinol binding protein 7, cellular |
| chr6_+_41458923 | 1.49 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr6_+_41392356 | 1.47 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr7_+_100178679 | 1.44 |
ENSMUST00000170954.2
ENSMUST00000049333.5 ENSMUST00000179842.1 |
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr10_+_112271123 | 1.42 |
ENSMUST00000092175.2
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr5_-_103977360 | 1.38 |
ENSMUST00000048118.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr7_+_44225430 | 1.38 |
ENSMUST00000075162.3
|
Klk1
|
kallikrein 1 |
| chr11_+_120530688 | 1.37 |
ENSMUST00000026119.7
|
Gcgr
|
glucagon receptor |
| chr11_-_4160286 | 1.37 |
ENSMUST00000093381.4
ENSMUST00000101626.2 |
Ccdc157
|
coiled-coil domain containing 157 |
| chrX_+_20703906 | 1.37 |
ENSMUST00000033383.2
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr15_-_75566608 | 1.36 |
ENSMUST00000163116.1
ENSMUST00000023241.5 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr17_+_21691860 | 1.34 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
| chr14_+_118787894 | 1.34 |
ENSMUST00000047761.6
ENSMUST00000071546.7 |
Cldn10
|
claudin 10 |
| chr15_+_98571004 | 1.34 |
ENSMUST00000023728.6
|
4930415O20Rik
|
RIKEN cDNA 4930415O20 gene |
| chr6_+_138141569 | 1.33 |
ENSMUST00000118091.1
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr7_-_131322292 | 1.32 |
ENSMUST00000046611.7
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr3_-_75270073 | 1.30 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr8_-_22125030 | 1.29 |
ENSMUST00000169834.1
|
Nek5
|
NIMA (never in mitosis gene a)-related expressed kinase 5 |
| chr7_+_97453204 | 1.29 |
ENSMUST00000050732.7
ENSMUST00000121987.1 |
Kctd14
|
potassium channel tetramerisation domain containing 14 |
| chr7_-_30924169 | 1.27 |
ENSMUST00000074671.6
|
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr14_+_103046977 | 1.26 |
ENSMUST00000022722.6
|
Irg1
|
immunoresponsive gene 1 |
| chr11_+_82045705 | 1.25 |
ENSMUST00000021011.2
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr4_+_120666562 | 1.25 |
ENSMUST00000094814.4
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr15_-_5063741 | 1.25 |
ENSMUST00000110689.3
|
C7
|
complement component 7 |
| chr19_-_34166037 | 1.24 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr3_-_20275659 | 1.24 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr1_+_90915064 | 1.23 |
ENSMUST00000027528.6
|
Mlph
|
melanophilin |
| chr7_+_140835018 | 1.22 |
ENSMUST00000106050.1
ENSMUST00000026554.4 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr16_+_96361749 | 1.22 |
ENSMUST00000000163.6
ENSMUST00000081093.3 ENSMUST00000113795.1 |
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr15_-_82338801 | 1.18 |
ENSMUST00000023088.7
|
Naga
|
N-acetyl galactosaminidase, alpha |
| chr3_-_122924103 | 1.17 |
ENSMUST00000180557.1
|
4933405D12Rik
|
RIKEN cDNA 4933405D12 gene |
| chr4_-_141846359 | 1.15 |
ENSMUST00000037059.10
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr3_+_19957240 | 1.15 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr6_+_96113146 | 1.14 |
ENSMUST00000122120.1
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr18_-_80247102 | 1.14 |
ENSMUST00000166219.1
|
Hsbp1l1
|
heat shock factor binding protein 1-like 1 |
| chr5_-_86906937 | 1.13 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr14_-_63245219 | 1.13 |
ENSMUST00000118022.1
ENSMUST00000067417.3 |
Gata4
|
GATA binding protein 4 |
| chr14_-_50795689 | 1.12 |
ENSMUST00000095932.3
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr11_+_70166623 | 1.12 |
ENSMUST00000102571.3
ENSMUST00000178945.1 ENSMUST00000000327.6 |
Clec10a
|
C-type lectin domain family 10, member A |
| chrX_+_101376359 | 1.11 |
ENSMUST00000119080.1
|
Gjb1
|
gap junction protein, beta 1 |
| chrX_-_108664891 | 1.10 |
ENSMUST00000178160.1
|
Gm379
|
predicted gene 379 |
| chr6_-_112489808 | 1.08 |
ENSMUST00000053306.6
|
Oxtr
|
oxytocin receptor |
| chr3_+_89229046 | 1.07 |
ENSMUST00000041142.3
|
Muc1
|
mucin 1, transmembrane |
| chr10_+_22158566 | 1.06 |
ENSMUST00000181645.1
ENSMUST00000105522.2 |
Raet1e
H60b
|
retinoic acid early transcript 1E histocompatibility 60b |
| chr6_-_5496296 | 1.05 |
ENSMUST00000019721.4
|
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr1_+_171419027 | 1.05 |
ENSMUST00000171362.1
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr9_-_48605147 | 1.04 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr1_+_107511416 | 1.03 |
ENSMUST00000009356.4
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr9_+_45117813 | 1.03 |
ENSMUST00000170998.1
ENSMUST00000093855.3 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr2_+_172345565 | 1.02 |
ENSMUST00000028995.4
|
Fam210b
|
family with sequence similarity 210, member B |
| chr5_-_103977326 | 1.01 |
ENSMUST00000120320.1
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_+_171155512 | 1.01 |
ENSMUST00000111334.1
|
Mpz
|
myelin protein zero |
| chr19_+_56287943 | 1.00 |
ENSMUST00000166049.1
|
Habp2
|
hyaluronic acid binding protein 2 |
| chrX_-_167209149 | 1.00 |
ENSMUST00000112176.1
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr3_+_19957088 | 0.99 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr16_-_22161450 | 0.99 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr5_-_103977404 | 0.98 |
ENSMUST00000112803.2
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_+_107511489 | 0.98 |
ENSMUST00000064916.2
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr7_-_45370559 | 0.96 |
ENSMUST00000003971.7
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr1_-_173333503 | 0.96 |
ENSMUST00000038227.4
|
Darc
|
Duffy blood group, chemokine receptor |
| chr16_-_42340595 | 0.96 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr9_+_88581036 | 0.95 |
ENSMUST00000164661.2
|
Trim43a
|
tripartite motif-containing 43A |
| chr11_+_94936224 | 0.95 |
ENSMUST00000001547.7
|
Col1a1
|
collagen, type I, alpha 1 |
| chr1_-_131276914 | 0.95 |
ENSMUST00000161764.1
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr11_+_98412461 | 0.95 |
ENSMUST00000058295.5
|
Erbb2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) |
| chr6_+_90550789 | 0.95 |
ENSMUST00000130418.1
ENSMUST00000032175.8 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr7_-_3502465 | 0.93 |
ENSMUST00000065703.7
|
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr16_+_44765732 | 0.93 |
ENSMUST00000057488.8
|
Cd200r1
|
CD200 receptor 1 |
| chr13_-_56296551 | 0.92 |
ENSMUST00000021970.9
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr7_-_101867391 | 0.92 |
ENSMUST00000106982.1
|
Folr1
|
folate receptor 1 (adult) |
| chr5_-_113800356 | 0.91 |
ENSMUST00000160374.1
ENSMUST00000067853.5 |
Tmem119
|
transmembrane protein 119 |
| chr4_-_103026709 | 0.90 |
ENSMUST00000084382.5
ENSMUST00000106869.2 |
Insl5
|
insulin-like 5 |
| chr9_+_46998931 | 0.88 |
ENSMUST00000178065.1
|
Gm4791
|
predicted gene 4791 |
| chr6_-_124769548 | 0.87 |
ENSMUST00000149652.1
ENSMUST00000112476.1 ENSMUST00000004378.8 |
Eno2
|
enolase 2, gamma neuronal |
| chr19_+_40612392 | 0.87 |
ENSMUST00000134063.1
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr1_+_88087802 | 0.86 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr13_-_99900645 | 0.86 |
ENSMUST00000022150.6
|
Cartpt
|
CART prepropeptide |
| chr16_-_21787796 | 0.86 |
ENSMUST00000023559.5
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
| chr6_-_85832082 | 0.86 |
ENSMUST00000032073.6
|
Nat8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr5_-_120887582 | 0.86 |
ENSMUST00000086368.5
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr10_+_115817247 | 0.85 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr11_-_120573253 | 0.85 |
ENSMUST00000026122.4
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr4_-_155653184 | 0.84 |
ENSMUST00000030937.1
|
Mmp23
|
matrix metallopeptidase 23 |
| chr16_-_23890805 | 0.84 |
ENSMUST00000004480.3
|
Sst
|
somatostatin |
| chr7_+_44207307 | 0.83 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr2_-_164356507 | 0.83 |
ENSMUST00000109367.3
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr9_+_92250039 | 0.83 |
ENSMUST00000093801.3
|
Plscr1
|
phospholipid scramblase 1 |
| chr18_-_53418004 | 0.82 |
ENSMUST00000025419.7
|
Ppic
|
peptidylprolyl isomerase C |
| chr5_-_120812506 | 0.82 |
ENSMUST00000117193.1
ENSMUST00000130045.1 |
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr4_-_4138817 | 0.80 |
ENSMUST00000133567.1
|
Penk
|
preproenkephalin |
| chr11_+_95010277 | 0.80 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr8_+_116504973 | 0.79 |
ENSMUST00000078170.5
|
Dynlrb2
|
dynein light chain roadblock-type 2 |
| chr9_+_5308828 | 0.79 |
ENSMUST00000162846.1
ENSMUST00000027012.7 |
Casp4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr13_-_113100971 | 0.79 |
ENSMUST00000023897.5
|
Gzma
|
granzyme A |
| chr5_-_137601043 | 0.79 |
ENSMUST00000037620.7
ENSMUST00000154708.1 |
Mospd3
|
motile sperm domain containing 3 |
| chr7_-_66427469 | 0.79 |
ENSMUST00000015278.7
|
Aldh1a3
|
aldehyde dehydrogenase family 1, subfamily A3 |
| chr11_+_96931387 | 0.79 |
ENSMUST00000107633.1
|
Prr15l
|
proline rich 15-like |
| chr4_+_152199805 | 0.79 |
ENSMUST00000105652.2
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr4_-_140617062 | 0.78 |
ENSMUST00000154979.1
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr7_+_43950614 | 0.78 |
ENSMUST00000072204.4
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr4_-_114908892 | 0.78 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr9_-_71592346 | 0.78 |
ENSMUST00000093823.1
|
Myzap
|
myocardial zonula adherens protein |
| chr17_-_34187219 | 0.78 |
ENSMUST00000173831.1
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr15_-_66560997 | 0.78 |
ENSMUST00000048372.5
|
Tmem71
|
transmembrane protein 71 |
| chr2_+_144556229 | 0.76 |
ENSMUST00000143573.1
ENSMUST00000028916.8 ENSMUST00000155258.1 |
Sec23b
|
SEC23B (S. cerevisiae) |
| chr4_+_120854786 | 0.76 |
ENSMUST00000071093.2
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr7_+_28437447 | 0.76 |
ENSMUST00000108292.2
ENSMUST00000108289.1 |
Gmfg
|
glia maturation factor, gamma |
| chr8_+_13435459 | 0.76 |
ENSMUST00000167071.1
ENSMUST00000167505.1 |
Tmem255b
|
transmembrane protein 255B |
| chr15_-_76307231 | 0.75 |
ENSMUST00000023222.6
ENSMUST00000164189.1 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr11_+_83703991 | 0.75 |
ENSMUST00000092836.5
|
Wfdc17
|
WAP four-disulfide core domain 17 |
| chr5_+_125003440 | 0.75 |
ENSMUST00000036109.3
|
Fam101a
|
family with sequence similarity 101, member A |
| chr14_+_65970804 | 0.75 |
ENSMUST00000138191.1
|
Clu
|
clusterin |
| chr10_-_62422427 | 0.74 |
ENSMUST00000020277.8
|
Hkdc1
|
hexokinase domain containing 1 |
| chr7_-_38227975 | 0.74 |
ENSMUST00000098513.4
|
Plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr7_+_107567445 | 0.73 |
ENSMUST00000120990.1
|
Olfml1
|
olfactomedin-like 1 |
| chr2_+_36049453 | 0.72 |
ENSMUST00000028256.4
|
Morn5
|
MORN repeat containing 5 |
| chr18_-_32139570 | 0.71 |
ENSMUST00000171765.1
|
Proc
|
protein C |
| chr8_+_75093591 | 0.71 |
ENSMUST00000005548.6
|
Hmox1
|
heme oxygenase (decycling) 1 |
| chr4_+_154170730 | 0.71 |
ENSMUST00000030897.8
|
Megf6
|
multiple EGF-like-domains 6 |
| chr2_-_80447625 | 0.71 |
ENSMUST00000028389.3
|
Frzb
|
frizzled-related protein |
| chr1_-_135375233 | 0.70 |
ENSMUST00000041240.3
|
Shisa4
|
shisa homolog 4 (Xenopus laevis) |
| chr18_-_37644185 | 0.70 |
ENSMUST00000066272.4
|
Taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr15_-_78305603 | 0.70 |
ENSMUST00000096356.3
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) |
| chr7_-_3845050 | 0.70 |
ENSMUST00000108615.3
ENSMUST00000119469.1 |
Pira2
|
paired-Ig-like receptor A2 |
| chr10_-_9675163 | 0.70 |
ENSMUST00000100070.2
|
Samd5
|
sterile alpha motif domain containing 5 |
| chr1_+_21240581 | 0.69 |
ENSMUST00000027067.8
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr17_-_47400584 | 0.68 |
ENSMUST00000059348.7
|
Guca1a
|
guanylate cyclase activator 1a (retina) |
| chr11_-_97884122 | 0.68 |
ENSMUST00000093939.3
|
Fbxo47
|
F-box protein 47 |
| chr12_+_84009481 | 0.68 |
ENSMUST00000168120.1
|
Acot1
|
acyl-CoA thioesterase 1 |
| chr11_+_69991633 | 0.68 |
ENSMUST00000108592.1
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chrX_-_68893053 | 0.68 |
ENSMUST00000074894.1
|
Gm6812
|
predicted gene 6812 |
| chr10_-_127070254 | 0.68 |
ENSMUST00000060991.4
|
Tspan31
|
tetraspanin 31 |
| chr11_-_99986593 | 0.68 |
ENSMUST00000105050.2
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr9_-_39604124 | 0.68 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr5_-_137610626 | 0.67 |
ENSMUST00000142675.1
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr11_+_78499087 | 0.67 |
ENSMUST00000017488.4
|
Vtn
|
vitronectin |
| chr16_-_18621366 | 0.67 |
ENSMUST00000051160.2
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr11_-_69695753 | 0.67 |
ENSMUST00000180946.1
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr15_-_75566811 | 0.67 |
ENSMUST00000065417.8
|
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr15_-_75567176 | 0.67 |
ENSMUST00000156032.1
ENSMUST00000127095.1 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr7_-_16221993 | 0.66 |
ENSMUST00000118795.1
ENSMUST00000119102.1 ENSMUST00000094816.2 ENSMUST00000163968.1 ENSMUST00000121123.1 |
Dhx34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr12_-_70231414 | 0.66 |
ENSMUST00000161083.1
|
Pygl
|
liver glycogen phosphorylase |
| chr7_+_4119525 | 0.66 |
ENSMUST00000119661.1
ENSMUST00000129423.1 |
Ttyh1
|
tweety homolog 1 (Drosophila) |
| chr12_-_12941827 | 0.66 |
ENSMUST00000043396.7
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr10_+_87859593 | 0.66 |
ENSMUST00000126490.1
|
Igf1
|
insulin-like growth factor 1 |
| chr4_-_55532453 | 0.66 |
ENSMUST00000132746.1
ENSMUST00000107619.2 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr15_+_76660564 | 0.66 |
ENSMUST00000004294.10
|
Kifc2
|
kinesin family member C2 |
| chr11_+_116593687 | 0.66 |
ENSMUST00000153476.1
|
Aanat
|
arylalkylamine N-acetyltransferase |
| chr11_-_69695802 | 0.65 |
ENSMUST00000108649.1
ENSMUST00000174159.1 ENSMUST00000181810.1 |
BC096441
Tnfsf12
|
cDNA sequence BC096441 tumor necrosis factor (ligand) superfamily, member 12 |
| chr9_+_64179289 | 0.65 |
ENSMUST00000034965.6
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr17_-_23684019 | 0.65 |
ENSMUST00000085989.5
|
Cldn9
|
claudin 9 |
| chr16_-_35871544 | 0.65 |
ENSMUST00000042665.8
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr1_+_37029307 | 0.65 |
ENSMUST00000067178.7
|
Vwa3b
|
von Willebrand factor A domain containing 3B |
| chr11_+_103133333 | 0.65 |
ENSMUST00000124928.1
ENSMUST00000062530.4 |
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr12_+_109734969 | 0.64 |
ENSMUST00000182268.1
ENSMUST00000181543.2 ENSMUST00000183116.1 |
Mirg
|
miRNA containing gene |
| chr10_+_67535465 | 0.64 |
ENSMUST00000145754.1
|
Egr2
|
early growth response 2 |
| chr9_-_58158498 | 0.63 |
ENSMUST00000168864.2
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr3_+_69222412 | 0.63 |
ENSMUST00000183126.1
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr7_+_29170204 | 0.63 |
ENSMUST00000098609.2
|
Ggn
|
gametogenetin |
| chr15_-_103366763 | 0.63 |
ENSMUST00000023128.6
|
Itga5
|
integrin alpha 5 (fibronectin receptor alpha) |
| chr5_+_90903864 | 0.63 |
ENSMUST00000075433.6
|
Cxcl2
|
chemokine (C-X-C motif) ligand 2 |
| chr1_+_88200601 | 0.62 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr8_+_120002720 | 0.62 |
ENSMUST00000108972.3
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0006571 | tyrosine biosynthetic process(GO:0006571) aromatic amino acid family biosynthetic process(GO:0009073) aromatic amino acid family biosynthetic process, prephenate pathway(GO:0009095) |
| 0.9 | 5.5 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.7 | 2.1 | GO:0035938 | androgen catabolic process(GO:0006710) estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.7 | 2.0 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.6 | 5.2 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.6 | 1.7 | GO:1905204 | cardiac muscle tissue regeneration(GO:0061026) negative regulation of connective tissue replacement(GO:1905204) |
| 0.5 | 9.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.5 | 0.5 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.4 | 3.6 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.4 | 1.4 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.7 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.3 | 1.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.3 | 0.3 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.3 | 0.9 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 0.6 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 0.3 | 1.4 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.3 | 0.6 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.3 | 0.9 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.3 | 3.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.1 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.3 | 0.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.3 | 1.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.3 | 1.1 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.3 | 1.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 1.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 0.8 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.3 | 0.3 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.2 | 0.7 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.2 | 0.9 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.2 | 2.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.2 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.2 | 0.7 | GO:1901376 | mycotoxin metabolic process(GO:0043385) mycotoxin catabolic process(GO:0043387) aflatoxin metabolic process(GO:0046222) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound metabolic process(GO:1901376) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.2 | 1.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 0.2 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.2 | 1.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.2 | 1.1 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.2 | 0.7 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 7.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.1 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 0.6 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 0.8 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 2.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 0.8 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 0.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.6 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.2 | 1.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 4.8 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 0.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.2 | 0.5 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.2 | 1.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 0.8 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.2 | 0.5 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 0.5 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 1.0 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 0.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 7.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 0.5 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.2 | 0.6 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 0.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 0.5 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 1.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 0.6 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.4 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 2.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.6 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.1 | 0.6 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.6 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.6 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.6 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 1.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.5 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 1.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.8 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.8 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.1 | 0.9 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.1 | 0.4 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.1 | 0.5 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.5 | GO:0071611 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 1.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.4 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.1 | 0.5 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 3.3 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 0.3 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.1 | 0.7 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.3 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.6 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.1 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 1.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 | 0.2 | GO:0014055 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.1 | 0.6 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 0.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.4 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.1 | 0.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.9 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.1 | 0.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.4 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.1 | 0.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.9 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.1 | 0.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 0.2 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 1.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:0060376 | development of secondary male sexual characteristics(GO:0046544) positive regulation of mast cell differentiation(GO:0060376) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 1.5 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.3 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 0.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.4 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 0.4 | GO:0072672 | neutrophil extravasation(GO:0072672) |
| 0.1 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.7 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 0.3 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 0.3 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 9.7 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 1.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.4 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.3 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 0.4 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.5 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.4 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.1 | 1.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.3 | GO:0045964 | negative regulation of serotonin secretion(GO:0014063) positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.4 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 0.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.3 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.5 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.1 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) regulation of estrogen secretion(GO:2000861) |
| 0.1 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.1 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.4 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0001802 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) positive regulation of type I hypersensitivity(GO:0001812) peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.7 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.1 | 0.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.2 | GO:1904430 | negative regulation of mitotic recombination(GO:0045950) negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.1 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.1 | 0.5 | GO:0065005 | protein-lipid complex assembly(GO:0065005) |
| 0.1 | 0.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.1 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.4 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.2 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 0.2 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.5 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0035546 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.6 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.3 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 1.2 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.2 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 1.3 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.9 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 | 0.3 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 1.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.2 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) regulation of cholesterol import(GO:0060620) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 2.0 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.8 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.1 | GO:0002584 | negative regulation of antigen processing and presentation(GO:0002578) negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.0 | 0.4 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 2.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.1 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) taste bud development(GO:0061193) |
| 0.0 | 1.9 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:0010574 | regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.0 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.4 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm morphogenesis(GO:0048319) axial mesoderm formation(GO:0048320) |
| 0.0 | 0.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 1.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.0 | GO:1902837 | amino acid import across plasma membrane(GO:0089718) amino acid import into cell(GO:1902837) |
| 0.0 | 0.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 3.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 1.2 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.1 | GO:2000617 | positive regulation of histone H3-K14 acetylation(GO:0071442) positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:2000158 | regulation of ubiquitin-specific protease activity(GO:2000152) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.6 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.3 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.6 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.1 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.2 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.2 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.4 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.0 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.0 | 0.8 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.7 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.2 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 | 1.2 | GO:2000116 | regulation of cysteine-type endopeptidase activity(GO:2000116) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 2.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 1.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.3 | 8.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 0.9 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.3 | 7.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 0.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 5.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 0.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 5.9 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.2 | 1.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.6 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.0 | GO:0042567 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 4.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.4 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 7.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 52.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0036452 | ESCRT I complex(GO:0000813) ESCRT complex(GO:0036452) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0030670 | external encapsulating structure(GO:0030312) phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 5.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.5 | 1.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 3.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 2.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.5 | 1.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.4 | 3.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 2.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 2.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 0.9 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 2.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 0.7 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.2 | 36.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 0.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 0.6 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 1.0 | GO:0008384 | NF-kappaB-inducing kinase activity(GO:0004704) IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 0.9 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.2 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 1.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 0.5 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 0.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.9 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 0.9 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 0.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 1.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.2 | 3.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 0.5 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.2 | 0.7 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 0.3 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 4.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.4 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.4 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 1.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 4.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 1.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.9 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.8 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.5 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 1.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 1.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 1.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.3 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.1 | 0.4 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 1.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.3 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.2 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.2 | GO:0070737 | protein-glycine ligase activity, elongating(GO:0070737) |
| 0.1 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 6.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 2.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 13.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.0 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 2.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.0 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 7.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 17.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 11.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 9.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.9 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 8.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 3.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 5.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.9 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 0.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.8 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 3.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |