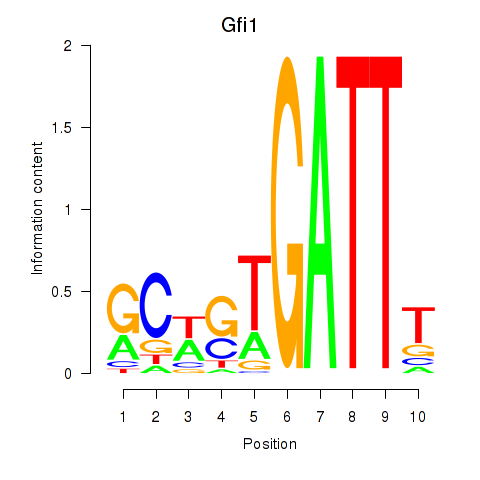
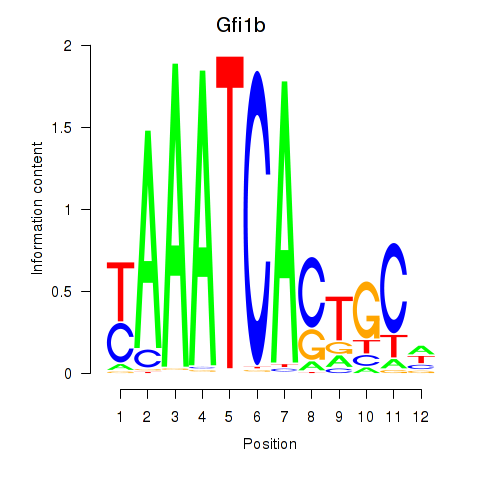
chr13_+_23947641
Show fit
5.89
ENSMUST00000055770.4
H1f1
H1.1 linker histone, cluster member
chr13_-_23946359
Show fit
5.69
ENSMUST00000091701.3
H3c1
H3 clustered histone 1
chr13_-_23806530
Show fit
3.57
ENSMUST00000062045.4
H1f4
H1.4 linker histone, cluster member
chr13_+_23755551
Show fit
3.33
ENSMUST00000079251.8
H2bc8
H2B clustered histone 8
chr13_+_21901791
Show fit
2.18
ENSMUST00000188775.2
H3c10
H3 clustered histone 10
chr13_-_23755374
Show fit
2.11
ENSMUST00000102969.6
H2ac8
H2A clustered histone 8
chr3_+_54268523
Show fit
1.59
ENSMUST00000117373.8
ENSMUST00000107985.10
ENSMUST00000073012.13
ENSMUST00000081564.13
Postn
periostin, osteoblast specific factor
chr13_-_21967540
Show fit
1.44
ENSMUST00000189457.2
H3c11
H3 clustered histone 11
chr12_-_99529767
Show fit
1.32
ENSMUST00000176928.3
ENSMUST00000223484.2
Foxn3
forkhead box N3
chr7_+_24408662
Show fit
1.25
ENSMUST00000206826.2
ENSMUST00000051714.9
Lypd10
Ly6/PLAUR domain containing 10
chr3_+_93427791
Show fit
1.25
ENSMUST00000029515.5
S100a11
S100 calcium binding protein A11
chr9_+_3000922
Show fit
1.19
ENSMUST00000151376.3
Gm10722
predicted gene 10722
chr9_+_3005126
Show fit
1.16
ENSMUST00000179881.2
Gm11168
predicted gene 11168
chr9_+_3013140
Show fit
1.14
ENSMUST00000143083.3
Gm10721
predicted gene 10721
chr10_+_127595590
Show fit
1.10
ENSMUST00000073639.6
Rdh1
retinol dehydrogenase 1 (all trans)
chr9_+_3027439
Show fit
1.09
ENSMUST00000179982.2
ENSMUST00000177875.8
Gm10717
predicted gene 10717
chr9_+_3023547
Show fit
1.07
ENSMUST00000099046.4
Gm10718
predicted gene 10718
chr9_+_3034599
Show fit
1.06
ENSMUST00000178641.2
Gm17535
predicted gene, 17535
chr9_+_3017408
Show fit
1.04
ENSMUST00000099049.4
Gm10719
predicted gene 10719
chr2_+_37101586
Show fit
0.96
ENSMUST00000214897.2
Olfr366
olfactory receptor 366
chr9_+_3025417
Show fit
0.95
ENSMUST00000075573.7
Gm10717
predicted gene 10717
chr9_+_3032001
Show fit
0.92
ENSMUST00000099056.4
ENSMUST00000179839.8
Gm10717
predicted gene 10717
chr1_-_140111138
Show fit
0.88
ENSMUST00000111976.9
ENSMUST00000066859.13
Cfh
complement component factor h
chr10_-_12745109
Show fit
0.76
ENSMUST00000218635.2
Utrn
utrophin
chr1_-_140111018
Show fit
0.75
ENSMUST00000192880.6
ENSMUST00000111977.8
Cfh
complement component factor h
chr5_-_66309244
Show fit
0.74
ENSMUST00000167950.8
Rbm47
RNA binding motif protein 47
chr9_+_3018753
Show fit
0.73
ENSMUST00000179272.2
Gm10719
predicted gene 10719
chr7_-_140597465
Show fit
0.71
ENSMUST00000211330.2
Ifitm6
interferon induced transmembrane protein 6
chr17_+_34311314
Show fit
0.69
ENSMUST00000025192.8
H2-Oa
histocompatibility 2, O region alpha locus
chr11_-_79421397
Show fit
0.68
ENSMUST00000103236.4
ENSMUST00000170799.8
ENSMUST00000170422.4
Evi2a
Evi2
ecotropic viral integration site 2a
chr9_+_3015654
Show fit
0.68
ENSMUST00000099050.4
Gm10720
predicted gene 10720
chr14_-_30850795
Show fit
0.66
ENSMUST00000049732.11
ENSMUST00000090205.5
ENSMUST00000064032.10
Smim4
small integral membrane protein 4
chr16_+_37909363
Show fit
0.65
ENSMUST00000023507.13
Gsk3b
glycogen synthase kinase 3 beta
chr14_-_33700719
Show fit
0.64
ENSMUST00000166737.2
Zfp488
zinc finger protein 488
chr13_+_47347301
Show fit
0.63
ENSMUST00000110111.4
Rnf144b
ring finger protein 144B
chr9_+_3004457
Show fit
0.63
ENSMUST00000178348.2
Gm11168
predicted gene 11168
chr17_-_43187280
Show fit
0.62
ENSMUST00000024709.9
ENSMUST00000233476.2
Cd2ap
CD2-associated protein
chr7_-_135318074
Show fit
0.62
ENSMUST00000033310.9
Mki67
antigen identified by monoclonal antibody Ki 67
chr19_+_11382092
Show fit
0.60
ENSMUST00000153546.8
Ms4a4c
membrane-spanning 4-domains, subfamily A, member 4C
chr2_-_89408791
Show fit
0.60
ENSMUST00000217402.2
Olfr1245
olfactory receptor 1245
chr7_+_23451695
Show fit
0.59
ENSMUST00000228331.2
Vmn1r174
vomeronasal 1 receptor 174
chr11_+_69804714
Show fit
0.56
ENSMUST00000072581.9
ENSMUST00000116358.8
Gps2
G protein pathway suppressor 2
chr3_+_124114504
Show fit
0.55
ENSMUST00000058994.6
Tram1l1
translocation associated membrane protein 1-like 1
chr1_+_17672117
Show fit
0.55
ENSMUST00000088476.4
Pi15
peptidase inhibitor 15
chr18_+_62681982
Show fit
0.55
ENSMUST00000055725.12
ENSMUST00000162365.8
Spink10
serine peptidase inhibitor, Kazal type 10
chrX_+_128650486
Show fit
0.54
ENSMUST00000167619.9
ENSMUST00000037854.15
Diaph2
diaphanous related formin 2
chr3_-_95725944
Show fit
0.54
ENSMUST00000200164.5
ENSMUST00000090791.8
ENSMUST00000197449.2
Rprd2
regulation of nuclear pre-mRNA domain containing 2
chr4_+_100633860
Show fit
0.53
ENSMUST00000030257.15
ENSMUST00000097955.3
Cachd1
cache domain containing 1
chr1_-_31261678
Show fit
0.52
ENSMUST00000187892.8
ENSMUST00000233331.2
4931428L18Rik
RIKEN cDNA 4931428L18 gene
chr2_-_160714473
Show fit
0.52
ENSMUST00000103111.9
Zhx3
zinc fingers and homeoboxes 3
chr10_+_127595639
Show fit
0.51
ENSMUST00000128247.2
Rdh16f1
RDH16 family member 1
chr6_+_6863269
Show fit
0.51
ENSMUST00000171311.8
ENSMUST00000160937.9
Dlx6
distal-less homeobox 6
chr8_+_56747613
Show fit
0.50
ENSMUST00000034026.10
Hpgd
hydroxyprostaglandin dehydrogenase 15 (NAD)
chr7_-_127490139
Show fit
0.50
ENSMUST00000205300.2
ENSMUST00000121394.3
Prss53
protease, serine 53
chr8_-_71257623
Show fit
0.50
ENSMUST00000212875.2
ENSMUST00000212001.2
ENSMUST00000212757.2
Mast3
microtubule associated serine/threonine kinase 3
chr12_-_99378855
Show fit
0.50
ENSMUST00000177269.2
Foxn3
forkhead box N3
chr9_+_7558449
Show fit
0.49
ENSMUST00000018765.4
Mmp8
matrix metallopeptidase 8
chr12_+_108145802
Show fit
0.49
ENSMUST00000221167.2
Ccnk
cyclin K
chr9_+_3036877
Show fit
0.49
ENSMUST00000155807.3
Gm10715
predicted gene 10715
chr6_-_66759611
Show fit
0.48
ENSMUST00000226457.2
Vmn1r38
vomeronasal 1 receptor 38
chr11_+_69805005
Show fit
0.48
ENSMUST00000057884.6
Gps2
G protein pathway suppressor 2
chr9_+_3037111
Show fit
0.47
ENSMUST00000177969.2
Gm10715
predicted gene 10715
chr12_-_111344139
Show fit
0.46
ENSMUST00000041965.5
Cdc42bpb
CDC42 binding protein kinase beta
chr9_+_110485561
Show fit
0.46
ENSMUST00000019803.9
ENSMUST00000196347.5
Ccdc12
coiled-coil domain containing 12
chr17_-_25234739
Show fit
0.45
ENSMUST00000233169.2
ENSMUST00000233774.2
Cramp1l
cramped chromatin regulator homolog 1
chr7_+_23400128
Show fit
0.45
ENSMUST00000226233.2
ENSMUST00000227987.2
Vmn1r173
vomeronasal 1 receptor 173
chr15_+_102368510
Show fit
0.45
ENSMUST00000164688.2
Prr13
proline rich 13
chr2_-_34990689
Show fit
0.44
ENSMUST00000226631.2
ENSMUST00000045776.5
ENSMUST00000226972.2
AI182371
expressed sequence AI182371
chr5_-_62923463
Show fit
0.44
ENSMUST00000076623.8
ENSMUST00000159470.3
Arap2
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
chr10_-_128505096
Show fit
0.44
ENSMUST00000238610.2
ENSMUST00000238712.2
Ikzf4
IKAROS family zinc finger 4
chr18_-_25887173
Show fit
0.43
ENSMUST00000225477.2
Celf4
CUGBP, Elav-like family member 4
chr6_+_40448286
Show fit
0.43
ENSMUST00000114779.9
Ssbp1
single-stranded DNA binding protein 1
chr11_-_23720953
Show fit
0.43
ENSMUST00000102864.5
Rel
reticuloendotheliosis oncogene
chrX_+_102400061
Show fit
0.43
ENSMUST00000116547.3
Chic1
cysteine-rich hydrophobic domain 1
chr18_-_61669641
Show fit
0.42
ENSMUST00000237557.2
ENSMUST00000171629.3
Arhgef37
Rho guanine nucleotide exchange factor (GEF) 37
chr2_+_164897547
Show fit
0.42
ENSMUST00000017799.12
ENSMUST00000073707.9
Cd40
CD40 antigen
chr12_+_76371634
Show fit
0.42
ENSMUST00000154078.3
ENSMUST00000095610.9
Akap5
A kinase (PRKA) anchor protein 5
chr9_+_86454018
Show fit
0.41
ENSMUST00000185566.7
ENSMUST00000034988.10
ENSMUST00000179212.3
Rwdd2a
RWD domain containing 2A
chr1_+_32211792
Show fit
0.40
ENSMUST00000027226.12
ENSMUST00000189878.2
ENSMUST00000188257.7
ENSMUST00000185666.2
Khdrbs2
KH domain containing, RNA binding, signal transduction associated 2
chr14_-_104081827
Show fit
0.40
ENSMUST00000022718.11
Ednrb
endothelin receptor type B
chr17_-_38618461
Show fit
0.40
ENSMUST00000213505.2
Olfr137
olfactory receptor 137
chr5_+_122781941
Show fit
0.40
ENSMUST00000100737.10
ENSMUST00000121489.8
ENSMUST00000031425.15
ENSMUST00000086247.6
P2rx7
purinergic receptor P2X, ligand-gated ion channel, 7
chr6_+_40448334
Show fit
0.40
ENSMUST00000031971.13
Ssbp1
single-stranded DNA binding protein 1
chr7_-_86016045
Show fit
0.40
ENSMUST00000213255.2
ENSMUST00000216700.2
ENSMUST00000213869.2
Olfr305
olfactory receptor 305
chr16_+_14523696
Show fit
0.39
ENSMUST00000023356.8
Snai2
snail family zinc finger 2
chrX_+_128650579
Show fit
0.39
ENSMUST00000113320.3
Diaph2
diaphanous related formin 2
chr14_+_24540745
Show fit
0.39
ENSMUST00000112384.10
Rps24
ribosomal protein S24
chr5_+_67765216
Show fit
0.38
ENSMUST00000087241.7
Shisa3
shisa family member 3
chr5_+_115983292
Show fit
0.38
ENSMUST00000137952.8
ENSMUST00000148245.8
Cit
citron
chr11_-_62348599
Show fit
0.38
ENSMUST00000127471.9
Ncor1
nuclear receptor co-repressor 1
chr9_+_39367997
Show fit
0.38
ENSMUST00000214818.3
Olfr954
olfactory receptor 954
chr8_+_105558204
Show fit
0.38
ENSMUST00000059449.7
Ces2b
carboxyesterase 2B
chr13_-_67508655
Show fit
0.38
ENSMUST00000081582.8
Zfp953
zinc finger protein 953
chr1_+_172525613
Show fit
0.37
ENSMUST00000038495.5
Crp
C-reactive protein, pentraxin-related
chr5_-_136003294
Show fit
0.37
ENSMUST00000154181.2
ENSMUST00000111152.8
ENSMUST00000111153.8
Ssc4d
scavenger receptor cysteine rich family, 4 domains
chr8_+_69333143
Show fit
0.37
ENSMUST00000015712.15
Lpl
lipoprotein lipase
chr7_+_6459167
Show fit
0.37
ENSMUST00000214301.3
Olfr1336
olfactory receptor 1336
chr1_-_78488795
Show fit
0.36
ENSMUST00000170511.3
BC035947
cDNA sequence BC035947
chr7_+_86645323
Show fit
0.36
ENSMUST00000233714.2
ENSMUST00000233648.2
ENSMUST00000164462.3
ENSMUST00000233730.2
Vmn2r79
vomeronasal 2, receptor 79
chr17_+_35354430
Show fit
0.36
ENSMUST00000173535.8
ENSMUST00000173952.8
Bag6
BCL2-associated athanogene 6
chr8_+_71207326
Show fit
0.36
ENSMUST00000110093.9
ENSMUST00000143118.3
ENSMUST00000034301.12
ENSMUST00000110090.8
Rab3a
RAB3A, member RAS oncogene family
chr16_-_55659194
Show fit
0.35
ENSMUST00000096026.9
ENSMUST00000036273.13
ENSMUST00000114457.8
Nfkbiz
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta
chr7_-_102998876
Show fit
0.35
ENSMUST00000215042.2
Olfr600
olfactory receptor 600
chr16_-_23339548
Show fit
0.35
ENSMUST00000089883.7
Masp1
mannan-binding lectin serine peptidase 1
chr7_+_101583283
Show fit
0.35
ENSMUST00000209639.2
ENSMUST00000210679.2
Numa1
nuclear mitotic apparatus protein 1
chr7_-_30259253
Show fit
0.35
ENSMUST00000108164.8
Lin37
lin-37 homolog (C. elegans)
chr5_-_113968483
Show fit
0.34
ENSMUST00000100874.6
Selplg
selectin, platelet (p-selectin) ligand
chr2_+_27776428
Show fit
0.34
ENSMUST00000028280.14
Col5a1
collagen, type V, alpha 1
chr8_+_117822593
Show fit
0.34
ENSMUST00000034308.16
ENSMUST00000176860.2
Bco1
beta-carotene oxygenase 1
chr10_-_129524028
Show fit
0.34
ENSMUST00000203785.3
ENSMUST00000217576.2
Olfr802
olfactory receptor 802
chr6_+_135339543
Show fit
0.34
ENSMUST00000205156.3
Emp1
epithelial membrane protein 1
chr2_+_111205867
Show fit
0.34
ENSMUST00000062407.6
Olfr1284
olfactory receptor 1284
chr17_-_35394971
Show fit
0.33
ENSMUST00000173324.8
Aif1
allograft inflammatory factor 1
chr2_+_37078210
Show fit
0.33
ENSMUST00000213969.2
Olfr365
olfactory receptor 365
chr3_+_90200470
Show fit
0.33
ENSMUST00000199754.5
Gatad2b
GATA zinc finger domain containing 2B
chr9_+_48073296
Show fit
0.33
ENSMUST00000216998.2
ENSMUST00000215780.2
Nxpe4
neurexophilin and PC-esterase domain family, member 4
chr8_+_70107131
Show fit
0.33
ENSMUST00000204285.3
Zfp964
zinc finger protein 964
chr15_-_76079891
Show fit
0.33
ENSMUST00000023226.13
Plec
plectin
chr16_+_65317389
Show fit
0.33
ENSMUST00000176330.8
ENSMUST00000004964.15
ENSMUST00000176038.8
Pou1f1
POU domain, class 1, transcription factor 1
chr9_-_121106209
Show fit
0.32
ENSMUST00000051479.13
ENSMUST00000171923.8
Ulk4
unc-51-like kinase 4
chr11_-_87783073
Show fit
0.32
ENSMUST00000213672.2
ENSMUST00000213928.2
Olfr462
olfactory receptor 462
chr14_-_50538979
Show fit
0.32
ENSMUST00000216195.2
ENSMUST00000214372.2
ENSMUST00000214756.2
Olfr733
olfactory receptor 733
chr5_-_124003553
Show fit
0.32
ENSMUST00000057145.7
Hcar2
hydroxycarboxylic acid receptor 2
chrX_+_71006577
Show fit
0.32
ENSMUST00000048790.7
Prrg3
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
chr8_-_85549452
Show fit
0.32
ENSMUST00000065539.6
Dand5
Links
DAN domain family member 5, BMP antagonist
chr19_+_56814700
Show fit
0.32
ENSMUST00000078723.11
ENSMUST00000121249.8
Tdrd1
tudor domain containing 1
chr6_+_129157576
Show fit
0.31
ENSMUST00000032260.6
Clec2d
C-type lectin domain family 2, member d
chr16_+_44914397
Show fit
0.31
ENSMUST00000061050.6
Ccdc80
coiled-coil domain containing 80
chr6_-_52185674
Show fit
0.31
ENSMUST00000062829.9
Hoxa6
homeobox A6
chr18_+_82928782
Show fit
0.31
ENSMUST00000235793.2
Zfp516
zinc finger protein 516
chr7_+_24920840
Show fit
0.31
ENSMUST00000055604.6
Zfp526
zinc finger protein 526
chr7_+_106462392
Show fit
0.31
ENSMUST00000080925.7
Olfr704
olfactory receptor 704
chr9_-_107512566
Show fit
0.31
ENSMUST00000055704.12
Gnai2
guanine nucleotide binding protein (G protein), alpha inhibiting 2
chr12_+_108145997
Show fit
0.30
ENSMUST00000101055.5
Ccnk
cyclin K
chr2_+_143757193
Show fit
0.29
ENSMUST00000103172.4
Dstn
destrin
chr7_-_80453033
Show fit
0.29
ENSMUST00000167377.3
Iqgap1
IQ motif containing GTPase activating protein 1
chr3_+_95434093
Show fit
0.29
ENSMUST00000015667.9
ENSMUST00000116304.3
Ctss
cathepsin S
chr12_-_32000534
Show fit
0.29
ENSMUST00000172314.9
Hbp1
high mobility group box transcription factor 1
chr2_-_88519531
Show fit
0.28
ENSMUST00000213545.2
Olfr1195
olfactory receptor 1195
chr10_-_81436671
Show fit
0.28
ENSMUST00000151858.8
ENSMUST00000142948.8
ENSMUST00000072020.9
Tle6
transducin-like enhancer of split 6
chr12_+_71062733
Show fit
0.28
ENSMUST00000046305.12
Arid4a
AT rich interactive domain 4A (RBP1-like)
chr1_+_133278248
Show fit
0.28
ENSMUST00000094556.3
Ren1
renin 1 structural
chr16_+_44913974
Show fit
0.28
ENSMUST00000099498.10
Ccdc80
coiled-coil domain containing 80
chr13_+_109397184
Show fit
0.28
ENSMUST00000153234.8
Pde4d
phosphodiesterase 4D, cAMP specific
chr19_+_6214416
Show fit
0.28
ENSMUST00000045042.8
ENSMUST00000237511.2
Batf2
basic leucine zipper transcription factor, ATF-like 2
chr10_-_62067026
Show fit
0.28
ENSMUST00000047883.11
Tspan15
tetraspanin 15
chr3_-_142101339
Show fit
0.27
ENSMUST00000198381.5
ENSMUST00000090134.12
ENSMUST00000196908.5
Pdlim5
PDZ and LIM domain 5
chrX_-_48980360
Show fit
0.27
ENSMUST00000217355.3
Olfr1322
olfactory receptor 1322
chr14_-_30850881
Show fit
0.27
ENSMUST00000203261.3
Smim4
small integral membrane protein 4
chr17_+_12338161
Show fit
0.27
ENSMUST00000024594.9
Agpat4
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
chr17_-_85995680
Show fit
0.25
ENSMUST00000024947.8
ENSMUST00000163568.4
Six2
sine oculis-related homeobox 2
chr10_-_93425553
Show fit
0.25
ENSMUST00000020203.7
Snrpf
small nuclear ribonucleoprotein polypeptide F
chrX_-_162810959
Show fit
0.25
ENSMUST00000033739.5
Car5b
carbonic anhydrase 5b, mitochondrial
chr6_+_15185202
Show fit
0.25
ENSMUST00000154448.2
Foxp2
forkhead box P2
chr7_-_44541787
Show fit
0.25
ENSMUST00000208551.2
ENSMUST00000208253.2
ENSMUST00000207654.2
ENSMUST00000207278.2
Med25
mediator complex subunit 25
chr4_+_109264284
Show fit
0.25
ENSMUST00000064129.14
ENSMUST00000106619.8
Ttc39a
tetratricopeptide repeat domain 39A
chr1_+_74582044
Show fit
0.24
ENSMUST00000113749.8
ENSMUST00000067916.13
ENSMUST00000113747.8
ENSMUST00000113750.8
Plcd4
phospholipase C, delta 4
chr18_+_61238714
Show fit
0.24
ENSMUST00000237706.2
Csf1r
colony stimulating factor 1 receptor
chr19_-_6134903
Show fit
0.24
ENSMUST00000160977.8
ENSMUST00000159859.2
ENSMUST00000025707.9
ENSMUST00000160712.8
ENSMUST00000237738.2
Zfpl1
zinc finger like protein 1
chr11_+_101207021
Show fit
0.24
ENSMUST00000142640.8
ENSMUST00000019470.14
Psme3
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki)
chr1_+_150268544
Show fit
0.24
ENSMUST00000124973.9
Tpr
translocated promoter region, nuclear basket protein
chr10_-_13428883
Show fit
0.24
ENSMUST00000019945.15
ENSMUST00000170376.8
Pex3
peroxisomal biogenesis factor 3
chr11_+_118319029
Show fit
0.24
ENSMUST00000124861.2
C1qtnf1
C1q and tumor necrosis factor related protein 1
chr7_-_104628324
Show fit
0.23
ENSMUST00000217091.2
ENSMUST00000210963.3
Olfr671
olfactory receptor 671
chr8_-_3798922
Show fit
0.23
ENSMUST00000208960.2
ENSMUST00000207979.2
Cd209a
CD209a antigen
chr3_+_59832635
Show fit
0.23
ENSMUST00000049476.3
Aadacl2fm1
AADACL2 family member 1
chr15_-_43733389
Show fit
0.23
ENSMUST00000067469.6
Tmem74
transmembrane protein 74
chrX_+_93768175
Show fit
0.23
ENSMUST00000101388.4
Zxdb
zinc finger, X-linked, duplicated B
chr8_-_3798941
Show fit
0.23
ENSMUST00000012847.3
Cd209a
CD209a antigen
chr7_-_137012444
Show fit
0.23
ENSMUST00000120340.2
ENSMUST00000117404.8
ENSMUST00000068996.13
9430038I01Rik
RIKEN cDNA 9430038I01 gene
chr3_+_121085373
Show fit
0.23
ENSMUST00000039442.12
Alg14
asparagine-linked glycosylation 14
chr7_-_44753168
Show fit
0.22
ENSMUST00000211085.2
ENSMUST00000210642.2
ENSMUST00000003512.9
Fcgrt
Fc fragment of IgG receptor and transporter
chr15_-_28025920
Show fit
0.22
ENSMUST00000090247.7
Trio
triple functional domain (PTPRF interacting)
chr11_+_70431063
Show fit
0.22
ENSMUST00000018429.12
ENSMUST00000108557.10
ENSMUST00000108556.2
Pld2
phospholipase D2
chr9_+_20492593
Show fit
0.22
ENSMUST00000115557.10
Zfp846
zinc finger protein 846
chr9_-_53447908
Show fit
0.22
ENSMUST00000150244.2
Atm
ataxia telangiectasia mutated
chr9_+_102885156
Show fit
0.22
ENSMUST00000035148.13
Slco2a1
solute carrier organic anion transporter family, member 2a1
chr19_+_6097111
Show fit
0.21
ENSMUST00000025723.9
Syvn1
synovial apoptosis inhibitor 1, synoviolin
chr12_+_71095112
Show fit
0.21
ENSMUST00000135709.2
Arid4a
AT rich interactive domain 4A (RBP1-like)
chr11_+_112673041
Show fit
0.21
ENSMUST00000000579.3
Sox9
SRY (sex determining region Y)-box 9
chr16_-_94046925
Show fit
0.21
ENSMUST00000228910.2
Hlcs
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase)
chr7_-_44393654
Show fit
0.21
ENSMUST00000149011.9
Zfp473
zinc finger protein 473
chr5_+_104350475
Show fit
0.21
ENSMUST00000066708.7
Dmp1
dentin matrix protein 1
chr5_+_102916637
Show fit
0.21
ENSMUST00000112852.8
Arhgap24
Rho GTPase activating protein 24
chr1_+_10108433
Show fit
0.21
ENSMUST00000071087.12
ENSMUST00000117415.8
Cspp1
centrosome and spindle pole associated protein 1
chrX_+_141009756
Show fit
0.20
ENSMUST00000112916.9
Nxt2
nuclear transport factor 2-like export factor 2
chr10_+_84412490
Show fit
0.20
ENSMUST00000020223.8
Tcp11l2
t-complex 11 (mouse) like 2
chr11_+_44508137
Show fit
0.20
ENSMUST00000109268.2
ENSMUST00000101326.10
ENSMUST00000081265.12
Ebf1
early B cell factor 1
chr5_-_123859070
Show fit
0.20
ENSMUST00000031376.12
Zcchc8
zinc finger, CCHC domain containing 8
chr7_-_80055168
Show fit
0.20
ENSMUST00000107362.10
ENSMUST00000135306.3
Furin
furin (paired basic amino acid cleaving enzyme)
chr5_-_114911548
Show fit
0.20
ENSMUST00000178440.8
ENSMUST00000043283.14
ENSMUST00000112185.9
ENSMUST00000155908.8
Git2
GIT ArfGAP 2
chr17_+_35354148
Show fit
0.19
ENSMUST00000166426.9
ENSMUST00000025250.14
Bag6
BCL2-associated athanogene 6
chr9_-_40442669
Show fit
0.19
ENSMUST00000119373.9
Gramd1b
GRAM domain containing 1B
chr7_+_106630381
Show fit
0.19
ENSMUST00000213623.2
Olfr713
olfactory receptor 713
chr17_+_36177498
Show fit
0.19
ENSMUST00000187690.7
ENSMUST00000113814.11
Ppp1r18
protein phosphatase 1, regulatory subunit 18
chr6_+_90078412
Show fit
0.19
ENSMUST00000089417.8
ENSMUST00000226577.2
Vmn1r50
vomeronasal 1 receptor 50
chr9_-_67336085
Show fit
0.18
ENSMUST00000213584.2
Tln2
talin 2
chr9_-_70328816
Show fit
0.18
ENSMUST00000034742.8
Ccnb2
cyclin B2
chr11_-_65679101
Show fit
0.18
ENSMUST00000152096.8
ENSMUST00000046963.10
Map2k4
mitogen-activated protein kinase kinase 4
chr13_-_100453124
Show fit
0.18
ENSMUST00000042220.3
Naip6
NLR family, apoptosis inhibitory protein 6