Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Id4
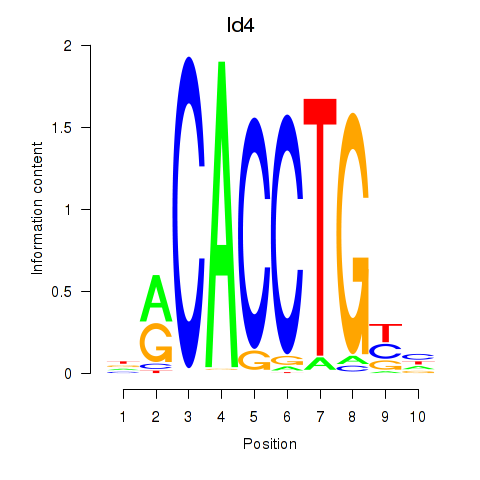
Z-value: 3.74
Transcription factors associated with Id4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Id4
|
ENSMUSG00000021379.3 | inhibitor of DNA binding 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Id4 | mm39_v1_chr13_+_48414582_48414704 | 0.98 | 4.3e-03 | Click! |
Activity profile of Id4 motif
Sorted Z-values of Id4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_79649683 | 4.28 |
ENSMUST00000162342.8
|
Ap1s3
|
adaptor-related protein complex AP-1, sigma 3 |
| chr15_-_101268036 | 3.11 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr19_+_8975249 | 2.83 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chrX_-_72913410 | 2.70 |
ENSMUST00000066576.12
ENSMUST00000114430.8 |
L1cam
|
L1 cell adhesion molecule |
| chr11_-_51647204 | 2.58 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr7_-_125681577 | 2.50 |
ENSMUST00000073935.7
|
Gsg1l
|
GSG1-like |
| chr13_+_47347301 | 2.46 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr7_-_29204812 | 2.30 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr10_-_41894360 | 2.17 |
ENSMUST00000162405.8
ENSMUST00000095729.11 ENSMUST00000161081.2 ENSMUST00000160262.9 |
Armc2
|
armadillo repeat containing 2 |
| chr1_+_131936022 | 2.10 |
ENSMUST00000146432.2
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr12_-_100995102 | 2.04 |
ENSMUST00000223097.2
|
Ccdc88c
|
coiled-coil domain containing 88C |
| chr2_+_71811526 | 1.97 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr15_-_72932853 | 1.91 |
ENSMUST00000170633.9
ENSMUST00000228960.2 |
Trappc9
|
trafficking protein particle complex 9 |
| chr6_-_83433357 | 1.88 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr13_-_12355604 | 1.88 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr7_+_3352019 | 1.86 |
ENSMUST00000100301.11
|
Prkcg
|
protein kinase C, gamma |
| chr4_-_135221810 | 1.85 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr5_-_25305621 | 1.83 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr9_-_123680726 | 1.79 |
ENSMUST00000084715.14
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr10_+_58649181 | 1.79 |
ENSMUST00000135526.9
ENSMUST00000153031.2 |
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr4_+_122889737 | 1.76 |
ENSMUST00000106252.9
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr9_-_57743989 | 1.74 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr11_-_120675009 | 1.74 |
ENSMUST00000026156.8
|
Rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr11_+_103061905 | 1.74 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr12_-_76756772 | 1.74 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr9_+_83430395 | 1.70 |
ENSMUST00000188030.2
|
Sh3bgrl2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr1_+_32211792 | 1.70 |
ENSMUST00000027226.12
ENSMUST00000189878.2 ENSMUST00000188257.7 ENSMUST00000185666.2 |
Khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr8_-_41087793 | 1.70 |
ENSMUST00000173957.2
ENSMUST00000048898.17 ENSMUST00000174205.8 |
Mtmr7
|
myotubularin related protein 7 |
| chr14_-_30075424 | 1.68 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr15_-_75438457 | 1.68 |
ENSMUST00000163116.8
ENSMUST00000023241.12 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr1_+_118555668 | 1.68 |
ENSMUST00000027629.10
|
Tfcp2l1
|
transcription factor CP2-like 1 |
| chr5_+_64969679 | 1.67 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr16_+_32090286 | 1.67 |
ENSMUST00000093183.5
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr2_-_168432111 | 1.65 |
ENSMUST00000029057.13
ENSMUST00000074618.10 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr5_-_125201872 | 1.65 |
ENSMUST00000055256.14
|
Ncor2
|
nuclear receptor co-repressor 2 |
| chr9_+_83430363 | 1.64 |
ENSMUST00000188241.7
ENSMUST00000113215.10 |
Sh3bgrl2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr3_-_122778052 | 1.63 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr11_-_103254257 | 1.61 |
ENSMUST00000092557.6
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr4_+_122889828 | 1.61 |
ENSMUST00000030407.8
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr5_+_117552042 | 1.60 |
ENSMUST00000180430.2
|
Ksr2
|
kinase suppressor of ras 2 |
| chr7_-_3828640 | 1.59 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr18_+_61058684 | 1.59 |
ENSMUST00000102888.10
ENSMUST00000025519.11 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr17_+_29077385 | 1.57 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr4_+_137004793 | 1.56 |
ENSMUST00000045747.5
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr5_-_62923463 | 1.56 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_125135434 | 1.52 |
ENSMUST00000134404.6
ENSMUST00000199561.2 |
Ncor2
|
nuclear receptor co-repressor 2 |
| chr19_+_10502612 | 1.52 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr8_-_70892204 | 1.51 |
ENSMUST00000076615.6
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr7_+_3339077 | 1.50 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_3339059 | 1.47 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr11_+_68986043 | 1.47 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr16_-_38162174 | 1.47 |
ENSMUST00000114740.3
|
Cfap91
|
cilia and flagella associated protein 91 |
| chr12_-_100995242 | 1.46 |
ENSMUST00000085096.10
|
Ccdc88c
|
coiled-coil domain containing 88C |
| chr5_-_66308421 | 1.46 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr1_-_39517761 | 1.46 |
ENSMUST00000193823.2
ENSMUST00000054462.11 |
Tbc1d8
|
TBC1 domain family, member 8 |
| chr11_+_6339442 | 1.45 |
ENSMUST00000109786.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr2_-_172300516 | 1.44 |
ENSMUST00000099060.2
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr15_-_103231921 | 1.44 |
ENSMUST00000229551.2
|
Zfp385a
|
zinc finger protein 385A |
| chr6_-_25689781 | 1.43 |
ENSMUST00000200812.2
|
Gpr37
|
G protein-coupled receptor 37 |
| chr1_+_59296065 | 1.42 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr7_+_96730915 | 1.42 |
ENSMUST00000206791.2
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr4_+_107825529 | 1.41 |
ENSMUST00000106713.5
ENSMUST00000238795.2 |
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr18_+_20691095 | 1.41 |
ENSMUST00000059787.15
ENSMUST00000120102.8 |
Dsg2
|
desmoglein 2 |
| chr5_+_99002293 | 1.40 |
ENSMUST00000031278.6
ENSMUST00000200388.2 |
Bmp3
|
bone morphogenetic protein 3 |
| chr14_+_26761023 | 1.38 |
ENSMUST00000223942.2
|
Il17rd
|
interleukin 17 receptor D |
| chr19_+_10502679 | 1.36 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr11_+_68322945 | 1.36 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chrX_+_162691978 | 1.36 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr7_+_44146029 | 1.33 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr10_+_60182630 | 1.32 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr7_+_44465714 | 1.31 |
ENSMUST00000208172.2
|
Nup62
|
nucleoporin 62 |
| chr8_+_70992334 | 1.30 |
ENSMUST00000093454.8
|
Ell
|
elongation factor RNA polymerase II |
| chr2_-_168432235 | 1.29 |
ENSMUST00000109184.8
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr11_-_109188917 | 1.28 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr1_+_61677977 | 1.28 |
ENSMUST00000075374.10
|
Pard3b
|
par-3 family cell polarity regulator beta |
| chr1_+_138891155 | 1.28 |
ENSMUST00000200533.5
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chrX_+_35375751 | 1.27 |
ENSMUST00000033418.8
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr3_-_100396635 | 1.26 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr11_+_6339330 | 1.25 |
ENSMUST00000012612.11
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr1_+_87254729 | 1.23 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr12_+_112688597 | 1.23 |
ENSMUST00000101018.11
ENSMUST00000092279.7 ENSMUST00000179041.8 ENSMUST00000222711.2 |
Cep170b
|
centrosomal protein 170B |
| chr1_-_123973223 | 1.22 |
ENSMUST00000112606.8
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr2_-_90410922 | 1.22 |
ENSMUST00000168621.3
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr11_-_74480870 | 1.21 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr12_-_100995305 | 1.20 |
ENSMUST00000068411.5
|
Ccdc88c
|
coiled-coil domain containing 88C |
| chr3_+_107008867 | 1.20 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr10_+_127595590 | 1.19 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr15_-_81074921 | 1.18 |
ENSMUST00000131235.9
ENSMUST00000134469.9 ENSMUST00000239114.2 ENSMUST00000149582.8 |
Mrtfa
|
myocardin related transcription factor A |
| chr10_-_128245501 | 1.18 |
ENSMUST00000172348.8
ENSMUST00000166608.8 ENSMUST00000164199.8 ENSMUST00000171370.2 ENSMUST00000026439.14 |
Nabp2
|
nucleic acid binding protein 2 |
| chr11_-_69088635 | 1.18 |
ENSMUST00000094078.4
ENSMUST00000021262.10 |
Alox8
|
arachidonate 8-lipoxygenase |
| chr8_+_66070661 | 1.17 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr11_-_69304501 | 1.16 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr7_-_12468931 | 1.16 |
ENSMUST00000233304.2
ENSMUST00000233373.2 ENSMUST00000233874.2 ENSMUST00000233902.2 |
Vmn2r56
|
vomeronasal 2, receptor 56 |
| chr12_+_84616536 | 1.15 |
ENSMUST00000021665.12
ENSMUST00000169934.4 |
Vsx2
|
visual system homeobox 2 |
| chr1_-_156767123 | 1.14 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_+_44893077 | 1.13 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr16_+_13074345 | 1.12 |
ENSMUST00000009713.14
ENSMUST00000115809.8 |
Mrtfb
|
myocardin related transcription factor B |
| chr9_+_102885156 | 1.12 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr14_-_47426863 | 1.12 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr6_+_82379456 | 1.11 |
ENSMUST00000032122.11
|
Tacr1
|
tachykinin receptor 1 |
| chr1_-_125839897 | 1.11 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr8_+_121264161 | 1.10 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr15_+_81469538 | 1.10 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr7_-_3723381 | 1.09 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr3_-_92514799 | 1.09 |
ENSMUST00000195278.2
|
2210017I01Rik
|
RIKEN cDNA 2210017I01 gene |
| chr6_-_124208815 | 1.08 |
ENSMUST00000233936.2
ENSMUST00000100968.4 |
Vmn2r27
|
vomeronasal 2, receptor27 |
| chr7_+_27879650 | 1.08 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr7_-_96981221 | 1.07 |
ENSMUST00000139582.9
|
Usp35
|
ubiquitin specific peptidase 35 |
| chr6_+_82379768 | 1.07 |
ENSMUST00000203775.2
|
Tacr1
|
tachykinin receptor 1 |
| chr12_+_112978051 | 1.07 |
ENSMUST00000223502.2
ENSMUST00000084891.5 ENSMUST00000220541.2 |
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr19_-_10970541 | 1.06 |
ENSMUST00000145110.3
ENSMUST00000144485.2 ENSMUST00000087923.4 |
Ms4a15
|
membrane-spanning 4-domains, subfamily A, member 15 |
| chr15_-_76084776 | 1.06 |
ENSMUST00000169108.8
ENSMUST00000170728.8 |
Plec
|
plectin |
| chr19_-_10502468 | 1.05 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chrX_+_167819606 | 1.04 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chrX_-_97934387 | 1.03 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr11_-_70578744 | 1.03 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr16_-_36695166 | 1.03 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chr6_+_47221293 | 1.03 |
ENSMUST00000199100.5
|
Cntnap2
|
contactin associated protein-like 2 |
| chr8_-_4267260 | 1.02 |
ENSMUST00000168386.9
|
Prr36
|
proline rich 36 |
| chr5_+_150119860 | 1.02 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr14_-_70873385 | 1.01 |
ENSMUST00000228295.2
ENSMUST00000022695.16 |
Dmtn
|
dematin actin binding protein |
| chr5_+_147366953 | 1.00 |
ENSMUST00000031651.15
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr1_+_132119169 | 1.00 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chrX_+_71006577 | 1.00 |
ENSMUST00000048790.7
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr7_-_24831892 | 0.99 |
ENSMUST00000108418.11
ENSMUST00000175774.9 ENSMUST00000108415.10 ENSMUST00000098679.10 ENSMUST00000108417.10 ENSMUST00000108416.10 ENSMUST00000108413.8 ENSMUST00000176408.8 |
Pou2f2
|
POU domain, class 2, transcription factor 2 |
| chr6_-_48422759 | 0.99 |
ENSMUST00000114561.9
|
Zfp467
|
zinc finger protein 467 |
| chr9_+_108174052 | 0.96 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr17_+_49735413 | 0.96 |
ENSMUST00000173033.8
|
Mocs1
|
molybdenum cofactor synthesis 1 |
| chr5_+_3978032 | 0.96 |
ENSMUST00000143365.8
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr1_+_78286946 | 0.95 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr2_-_33261411 | 0.95 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr1_-_138775317 | 0.95 |
ENSMUST00000093486.10
ENSMUST00000046870.13 |
Lhx9
|
LIM homeobox protein 9 |
| chr10_-_81037878 | 0.94 |
ENSMUST00000005069.8
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr17_+_47747657 | 0.94 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr6_+_3993774 | 0.94 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr9_-_51076724 | 0.93 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr2_-_172782089 | 0.93 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr1_-_88133472 | 0.92 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr16_+_16887991 | 0.92 |
ENSMUST00000232258.2
|
Ypel1
|
yippee like 1 |
| chr15_-_76082346 | 0.92 |
ENSMUST00000072692.11
|
Plec
|
plectin |
| chr15_-_71906051 | 0.92 |
ENSMUST00000159993.8
|
Col22a1
|
collagen, type XXII, alpha 1 |
| chr16_+_95058417 | 0.91 |
ENSMUST00000113861.8
ENSMUST00000113854.8 ENSMUST00000113862.8 ENSMUST00000037154.14 ENSMUST00000113855.8 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr6_-_48422612 | 0.91 |
ENSMUST00000114556.2
|
Zfp467
|
zinc finger protein 467 |
| chr6_+_72074545 | 0.90 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr7_-_30672824 | 0.90 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr14_-_104081119 | 0.89 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr6_-_24664959 | 0.89 |
ENSMUST00000041737.8
ENSMUST00000031695.15 |
Wasl
|
WASP like actin nucleation promoting factor |
| chr11_-_94133527 | 0.89 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr8_+_68729219 | 0.88 |
ENSMUST00000066594.4
|
Sh2d4a
|
SH2 domain containing 4A |
| chr1_-_128520002 | 0.88 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr3_+_20043315 | 0.88 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin |
| chr10_-_127016448 | 0.88 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chr1_-_52766615 | 0.88 |
ENSMUST00000156876.8
ENSMUST00000087701.4 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr7_+_24982206 | 0.88 |
ENSMUST00000165239.3
|
Cic
|
capicua transcriptional repressor |
| chr17_-_32408431 | 0.88 |
ENSMUST00000087721.10
ENSMUST00000162117.3 |
Ephx3
|
epoxide hydrolase 3 |
| chrM_+_2743 | 0.87 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr18_-_35760260 | 0.87 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr7_-_80051455 | 0.87 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr7_+_4140031 | 0.87 |
ENSMUST00000128756.8
ENSMUST00000132086.8 ENSMUST00000037472.13 ENSMUST00000117274.8 ENSMUST00000121270.8 ENSMUST00000144248.3 |
Leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr10_+_74802996 | 0.85 |
ENSMUST00000037813.5
|
Gnaz
|
guanine nucleotide binding protein, alpha z subunit |
| chr11_+_6339061 | 0.85 |
ENSMUST00000109787.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr17_-_43187280 | 0.85 |
ENSMUST00000024709.9
ENSMUST00000233476.2 |
Cd2ap
|
CD2-associated protein |
| chr6_-_34887530 | 0.85 |
ENSMUST00000149448.8
ENSMUST00000133336.8 |
Wdr91
|
WD repeat domain 91 |
| chr16_-_55642491 | 0.85 |
ENSMUST00000114458.8
|
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr11_+_33913013 | 0.84 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chrX_-_132882514 | 0.84 |
ENSMUST00000113297.9
ENSMUST00000174542.2 ENSMUST00000033608.15 ENSMUST00000113294.8 |
Sytl4
|
synaptotagmin-like 4 |
| chr9_+_46910039 | 0.84 |
ENSMUST00000178065.3
|
Gm4791
|
predicted gene 4791 |
| chr6_-_48422307 | 0.84 |
ENSMUST00000114563.8
ENSMUST00000114558.8 ENSMUST00000101443.10 |
Zfp467
|
zinc finger protein 467 |
| chr2_-_93164812 | 0.83 |
ENSMUST00000111265.9
|
Tspan18
|
tetraspanin 18 |
| chr1_-_105284407 | 0.83 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr14_-_21102487 | 0.83 |
ENSMUST00000154460.8
ENSMUST00000130291.8 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr7_+_123582021 | 0.83 |
ENSMUST00000106437.2
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr10_-_80096842 | 0.82 |
ENSMUST00000105363.8
|
Gamt
|
guanidinoacetate methyltransferase |
| chr13_-_58261406 | 0.82 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3 |
| chr7_+_6346723 | 0.82 |
ENSMUST00000207173.3
|
Gm3854
|
predicted gene 3854 |
| chr13_+_41759380 | 0.82 |
ENSMUST00000221694.2
|
Tmem170b
|
transmembrane protein 170B |
| chr17_-_71309012 | 0.82 |
ENSMUST00000128179.2
ENSMUST00000150456.2 ENSMUST00000233357.2 ENSMUST00000233417.2 |
Myl12a
Gm49909
|
myosin, light chain 12A, regulatory, non-sarcomeric predicted gene, 49909 |
| chr11_+_96820091 | 0.82 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr2_-_29983618 | 0.81 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr15_-_76004395 | 0.81 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr17_+_35455532 | 0.81 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr1_-_105284383 | 0.81 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr17_-_47143214 | 0.81 |
ENSMUST00000233537.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr11_+_68447012 | 0.81 |
ENSMUST00000053211.8
|
Mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr17_-_23964807 | 0.80 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr2_-_33321306 | 0.79 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr7_+_24990596 | 0.79 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr8_+_56747613 | 0.79 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr1_-_184615415 | 0.79 |
ENSMUST00000048308.6
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr5_-_66308666 | 0.78 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr11_+_102652228 | 0.78 |
ENSMUST00000103081.10
ENSMUST00000068150.7 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr12_-_34956910 | 0.77 |
ENSMUST00000239321.2
|
Hdac9
|
histone deacetylase 9 |
| chr4_-_135300934 | 0.77 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr2_+_32253016 | 0.77 |
ENSMUST00000132028.8
ENSMUST00000136079.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr7_-_127324788 | 0.77 |
ENSMUST00000076091.4
|
Ctf2
|
cardiotrophin 2 |
| chr7_+_4140474 | 0.77 |
ENSMUST00000154571.7
|
Leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chrX_+_23559282 | 0.77 |
ENSMUST00000035766.13
ENSMUST00000101670.3 |
Wdr44
|
WD repeat domain 44 |
| chr5_+_137639538 | 0.77 |
ENSMUST00000177466.8
ENSMUST00000166099.3 |
Sap25
|
sin3 associated polypeptide |
| chr13_+_25240138 | 0.77 |
ENSMUST00000069614.7
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr12_+_3856510 | 0.77 |
ENSMUST00000172719.8
|
Dnmt3a
|
DNA methyltransferase 3A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Id4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.6 | 1.9 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.6 | 1.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.5 | 1.6 | GO:0072034 | positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) negative regulation of male gonad development(GO:2000019) |
| 0.5 | 1.5 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.5 | 2.5 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.5 | 1.5 | GO:2000387 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.5 | 1.4 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.4 | 0.8 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.4 | 1.9 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.4 | 1.9 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 1.1 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.4 | 1.5 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.4 | 1.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.4 | 2.1 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.4 | 2.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 0.7 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.3 | 1.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.3 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.3 | 1.0 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.3 | 1.3 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.3 | 1.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.3 | 1.9 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 0.9 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 0.9 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 0.9 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.3 | 0.9 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 1.8 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.3 | 1.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 1.7 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 0.8 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.3 | 1.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.3 | 3.5 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.3 | 1.3 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.3 | 0.3 | GO:0043585 | nose morphogenesis(GO:0043585) |
| 0.3 | 2.6 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 1.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 1.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 0.7 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.2 | 0.7 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 0.9 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.2 | 1.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 2.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.6 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.2 | 1.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 0.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.6 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 0.6 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 2.7 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 0.6 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 | 0.9 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 1.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 1.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.1 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 0.7 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.2 | 0.9 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 1.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.9 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.7 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 2.5 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.2 | 1.0 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 0.5 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 0.7 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 1.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 0.6 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.2 | 0.6 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.2 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.2 | 1.4 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.2 | 0.5 | GO:2000184 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.2 | 0.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.6 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.4 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.1 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.8 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 1.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.7 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 2.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.4 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 2.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.8 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 1.8 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 1.7 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 1.0 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 1.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.4 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 1.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.5 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.8 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 2.7 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 1.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 1.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 1.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.5 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.2 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.1 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.5 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.4 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.1 | 1.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.6 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 1.7 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 4.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 0.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.6 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.4 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 3.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.1 | GO:1904000 | positive regulation of eating behavior(GO:1904000) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.8 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.2 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 1.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.8 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.2 | GO:0002930 | trabecular meshwork development(GO:0002930) dibenzo-p-dioxin metabolic process(GO:0018894) endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 2.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 1.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.4 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.6 | GO:0070535 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.2 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.6 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 2.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.5 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.2 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.3 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 1.5 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 1.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.3 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 1.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.5 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 1.0 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 1.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 1.2 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.0 | 3.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.4 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.3 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.0 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 1.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.2 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.4 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 1.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.6 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 2.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.6 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 2.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 1.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.7 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.5 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.4 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 1.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 1.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.4 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:0050908 | detection of visible light(GO:0009584) detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.3 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.7 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.7 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 2.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.1 | GO:0072014 | proximal tubule development(GO:0072014) regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 2.3 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.8 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.4 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.2 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0001963 | synaptic transmission, dopaminergic(GO:0001963) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 1.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.1 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.0 | 0.0 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.7 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0034033 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.2 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.0 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.0 | 0.0 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.0 | 0.1 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.5 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) growth plate cartilage chondrocyte proliferation(GO:0003419) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 1.6 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.4 | 2.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 1.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 1.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.3 | 1.3 | GO:0035363 | histone locus body(GO:0035363) |
| 0.2 | 1.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 1.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 1.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 1.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.6 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.2 | 1.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 0.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 3.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 0.5 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.2 | 0.5 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 1.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 2.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 2.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.6 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.3 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 4.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 1.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 3.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 2.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 7.0 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 5.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 4.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 5.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.6 | 1.7 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.5 | 2.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.5 | 1.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 2.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 1.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 2.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 1.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 0.9 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.3 | 1.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 0.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 2.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 0.7 | GO:1902271 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.2 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 1.7 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 1.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 0.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 1.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 0.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 0.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 1.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 1.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 2.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.6 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 2.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 4.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.6 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.1 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 3.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.6 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 1.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 1.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.4 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.8 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.0 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 2.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 3.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 2.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 1.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 4.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 2.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 10.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 2.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 3.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.0 | 0.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 7.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 5.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 4.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 4.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 0.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 4.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 4.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 3.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 2.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.7 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |