Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
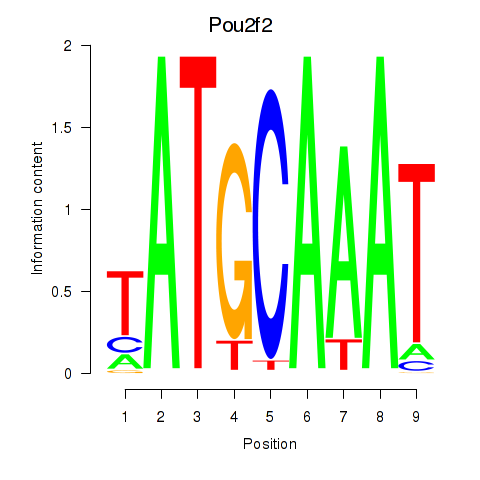
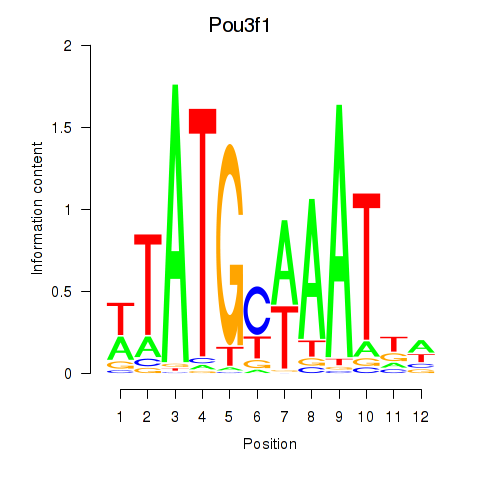
Results for Pou2f2_Pou3f1
Z-value: 7.07
Transcription factors associated with Pou2f2_Pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f2
|
ENSMUSG00000008496.20 | POU domain, class 2, transcription factor 2 |
|
Pou3f1
|
ENSMUSG00000090125.4 | POU domain, class 3, transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f1 | mm39_v1_chr4_+_124550600_124550600 | 0.95 | 1.2e-02 | Click! |
| Pou2f2 | mm39_v1_chr7_-_24831892_24831937 | 0.62 | 2.7e-01 | Click! |
Activity profile of Pou2f2_Pou3f1 motif
Sorted Z-values of Pou2f2_Pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23930717 | 38.78 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr13_-_23758370 | 20.98 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7 |
| chr3_-_96128196 | 20.35 |
ENSMUST00000090782.4
|
H2ac20
|
H2A clustered histone 20 |
| chr3_+_96177010 | 19.31 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr13_+_23868175 | 19.02 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr13_+_22220000 | 18.13 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr3_+_96152813 | 18.07 |
ENSMUST00000078756.7
ENSMUST00000090779.4 |
H2ac18
Gm20634
|
H2A clustered histone 18 predicted gene 20634 |
| chr13_+_21906214 | 17.73 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr13_+_22227359 | 17.63 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr3_-_96147592 | 17.39 |
ENSMUST00000074976.8
|
H2ac19
|
H2A clustered histone 19 |
| chr13_+_23755551 | 14.80 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr13_+_23728222 | 14.70 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr13_-_23867924 | 14.08 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr13_+_23935088 | 13.00 |
ENSMUST00000078369.3
|
H2ac4
|
H2A clustered histone 4 |
| chr13_-_24118139 | 12.46 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr13_-_23755374 | 11.25 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr13_-_21994366 | 10.33 |
ENSMUST00000091749.4
|
H2bc23
|
H2B clustered histone 23 |
| chr11_+_58845502 | 10.33 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr13_-_23805077 | 8.80 |
ENSMUST00000091704.7
ENSMUST00000051091.5 |
H2bc6
|
H2B clustered histone 6 |
| chr13_+_22017906 | 8.71 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr13_-_22219738 | 8.65 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr13_-_22227114 | 8.59 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr13_+_21971631 | 8.02 |
ENSMUST00000110473.3
ENSMUST00000102982.2 |
H2bc22
|
H2B clustered histone 22 |
| chr13_+_23718038 | 7.24 |
ENSMUST00000073261.3
|
H2ac10
|
H2A clustered histone 10 |
| chr13_+_21994588 | 6.43 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23 |
| chr13_+_23758555 | 6.16 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr11_+_58839716 | 5.90 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chr7_+_44667377 | 5.77 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr6_+_136495818 | 5.59 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr9_+_106099797 | 5.11 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr8_+_69333143 | 4.89 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr8_+_109441276 | 4.63 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr14_-_67106037 | 4.32 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr2_-_18053595 | 4.27 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr13_-_21971388 | 4.21 |
ENSMUST00000091751.3
|
H2ac22
|
H2A clustered histone 22 |
| chr17_-_35383867 | 4.06 |
ENSMUST00000025253.12
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr13_-_22017677 | 3.99 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr6_+_136495784 | 3.94 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_+_6552270 | 3.82 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr8_+_91635192 | 3.51 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr11_+_34264757 | 3.43 |
ENSMUST00000165963.9
ENSMUST00000093192.4 |
Insyn2b
|
inhibitory synaptic factor family member 2B |
| chr3_+_96127174 | 3.18 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr5_+_42225303 | 3.15 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr9_-_51076724 | 3.12 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr8_+_108669276 | 3.04 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr10_-_12745109 | 2.88 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr16_-_4698148 | 2.82 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr5_+_105879914 | 2.70 |
ENSMUST00000154807.2
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr13_+_41071077 | 2.51 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr4_-_128699838 | 2.45 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chrX_-_165992311 | 2.39 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr16_-_92494203 | 2.39 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr12_-_114752425 | 2.22 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr13_-_40887244 | 2.18 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr11_-_110142565 | 2.07 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr16_-_92622972 | 1.90 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr7_+_126376099 | 1.89 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr10_+_66932235 | 1.87 |
ENSMUST00000174317.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chrX_-_16777913 | 1.75 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr13_-_95661726 | 1.73 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr13_-_108026590 | 1.66 |
ENSMUST00000225822.4
ENSMUST00000225197.3 |
Zswim6
|
zinc finger SWIM-type containing 6 |
| chr9_-_85209340 | 1.65 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr7_+_24981604 | 1.62 |
ENSMUST00000163320.8
ENSMUST00000005578.13 |
Cic
|
capicua transcriptional repressor |
| chr6_-_99073156 | 1.62 |
ENSMUST00000175886.8
|
Foxp1
|
forkhead box P1 |
| chr10_+_128542120 | 1.61 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chr11_-_23720953 | 1.51 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr2_+_118877610 | 1.49 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr5_+_135135735 | 1.41 |
ENSMUST00000201977.4
ENSMUST00000005507.10 |
Mlxipl
|
MLX interacting protein-like |
| chr9_-_85209162 | 1.40 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr13_+_16186410 | 1.38 |
ENSMUST00000042603.14
|
Inhba
|
inhibin beta-A |
| chr11_-_5787743 | 1.37 |
ENSMUST00000109837.8
|
Polm
|
polymerase (DNA directed), mu |
| chr9_+_75682637 | 1.33 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chr11_+_6339061 | 1.32 |
ENSMUST00000109787.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr2_-_30364219 | 1.31 |
ENSMUST00000065134.4
|
Ier5l
|
immediate early response 5-like |
| chr1_-_66856695 | 1.30 |
ENSMUST00000068168.10
ENSMUST00000113987.2 |
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr18_-_60724855 | 1.30 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr2_+_90927053 | 1.30 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr5_+_53747796 | 1.30 |
ENSMUST00000113865.5
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr19_-_40260060 | 1.29 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr6_+_67586695 | 1.24 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr11_+_115272732 | 1.10 |
ENSMUST00000053288.6
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr19_+_53128901 | 1.09 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr4_-_152561896 | 1.07 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr5_-_135518098 | 1.06 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr2_-_35995283 | 1.06 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr8_+_121215155 | 1.00 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr4_-_129590372 | 1.00 |
ENSMUST00000137640.3
|
Tmem39b
|
transmembrane protein 39b |
| chr7_+_18962301 | 0.98 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr11_+_6339442 | 0.97 |
ENSMUST00000109786.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr7_+_121888520 | 0.93 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr17_+_47451868 | 0.92 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr11_+_6339330 | 0.92 |
ENSMUST00000012612.11
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr13_+_37529184 | 0.91 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr9_+_77959206 | 0.91 |
ENSMUST00000024104.9
|
Gcm1
|
glial cells missing homolog 1 |
| chr10_-_12490424 | 0.90 |
ENSMUST00000217994.2
|
Utrn
|
utrophin |
| chr5_-_124003553 | 0.90 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr2_+_118877594 | 0.88 |
ENSMUST00000152380.8
ENSMUST00000099542.9 |
Knl1
|
kinetochore scaffold 1 |
| chr13_+_102830029 | 0.88 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr5_-_21906330 | 0.86 |
ENSMUST00000115217.8
ENSMUST00000060899.9 |
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr7_-_102507962 | 0.82 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr6_+_68402550 | 0.81 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr7_-_28071919 | 0.80 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr16_+_42727926 | 0.80 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_-_63928339 | 0.79 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr16_+_44913974 | 0.78 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr12_-_114710326 | 0.78 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr12_+_55211069 | 0.76 |
ENSMUST00000218889.2
|
Srp54b
|
signal recognition particle 54B |
| chr12_-_114502585 | 0.76 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr7_+_140774962 | 0.75 |
ENSMUST00000047093.11
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr19_-_40260286 | 0.75 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr19_+_53128861 | 0.75 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr12_-_114451189 | 0.74 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr10_+_67373691 | 0.73 |
ENSMUST00000048289.14
ENSMUST00000130933.2 ENSMUST00000105438.9 ENSMUST00000146986.2 |
Egr2
|
early growth response 2 |
| chr12_-_113625906 | 0.73 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr13_+_102830104 | 0.70 |
ENSMUST00000172138.2
|
Cd180
|
CD180 antigen |
| chr11_+_69217078 | 0.70 |
ENSMUST00000018614.3
|
Kcnab3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr8_+_71069476 | 0.69 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr3_-_144525255 | 0.67 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr13_+_83723255 | 0.67 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr7_+_92390811 | 0.66 |
ENSMUST00000032879.15
|
Rab30
|
RAB30, member RAS oncogene family |
| chr5_+_123153072 | 0.66 |
ENSMUST00000051016.5
ENSMUST00000121652.8 |
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr5_+_53747556 | 0.65 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_164664920 | 0.65 |
ENSMUST00000132282.2
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr1_+_93163553 | 0.65 |
ENSMUST00000062202.14
|
Sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr2_-_57942844 | 0.64 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr8_+_71151581 | 0.63 |
ENSMUST00000095267.8
|
Jund
|
jun D proto-oncogene |
| chr18_+_20509771 | 0.63 |
ENSMUST00000076737.7
|
Dsg1b
|
desmoglein 1 beta |
| chr2_-_32977182 | 0.61 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr6_+_68279392 | 0.60 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chrX_-_73129195 | 0.60 |
ENSMUST00000140399.3
ENSMUST00000123362.8 ENSMUST00000100750.10 ENSMUST00000033770.13 |
Mecp2
|
methyl CpG binding protein 2 |
| chr3_-_80710097 | 0.60 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr9_+_65831489 | 0.57 |
ENSMUST00000130798.3
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr12_-_115916604 | 0.56 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr15_+_57558048 | 0.56 |
ENSMUST00000096430.11
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr17_-_24752683 | 0.56 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr4_-_123458465 | 0.56 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr12_-_56583582 | 0.55 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr1_-_163141230 | 0.54 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chrX_+_7744535 | 0.53 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr6_+_70675416 | 0.52 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr16_-_89615225 | 0.52 |
ENSMUST00000164263.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr6_-_72498676 | 0.51 |
ENSMUST00000234615.2
|
Sh2d6
|
SH2 domain containing 6 |
| chr12_-_114443071 | 0.50 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr7_-_44702269 | 0.46 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chr2_-_26393826 | 0.46 |
ENSMUST00000028288.5
|
Notch1
|
notch 1 |
| chr13_+_38529062 | 0.45 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr17_+_27775471 | 0.45 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr14_-_103220066 | 0.45 |
ENSMUST00000184744.2
|
Kctd12
|
potassium channel tetramerisation domain containing 12 |
| chr11_+_60360165 | 0.45 |
ENSMUST00000071880.9
ENSMUST00000081823.12 ENSMUST00000094135.9 |
Myo15
|
myosin XV |
| chr7_+_5018453 | 0.45 |
ENSMUST00000086349.5
ENSMUST00000207050.2 |
Zfp524
Gm44973
|
zinc finger protein 524 predicted gene 44973 |
| chr5_-_5315968 | 0.44 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr18_+_69654572 | 0.44 |
ENSMUST00000200862.4
|
Tcf4
|
transcription factor 4 |
| chr2_-_25982160 | 0.44 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr10_-_129751466 | 0.43 |
ENSMUST00000213438.2
|
Olfr816
|
olfactory receptor 816 |
| chr10_+_79763164 | 0.43 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr12_-_17374704 | 0.42 |
ENSMUST00000020884.16
ENSMUST00000095820.12 ENSMUST00000221129.2 ENSMUST00000127185.8 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr11_+_108814007 | 0.42 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr2_-_168048984 | 0.42 |
ENSMUST00000088001.6
|
Adnp
|
activity-dependent neuroprotective protein |
| chr1_-_163141278 | 0.41 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr4_+_97660971 | 0.41 |
ENSMUST00000152023.8
|
Nfia
|
nuclear factor I/A |
| chr5_-_86475595 | 0.40 |
ENSMUST00000122377.2
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr7_-_28072022 | 0.40 |
ENSMUST00000144700.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr1_-_14380418 | 0.37 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr12_-_115832846 | 0.35 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr16_+_91526169 | 0.34 |
ENSMUST00000114001.8
ENSMUST00000113999.8 ENSMUST00000064797.12 ENSMUST00000114002.9 ENSMUST00000095909.10 ENSMUST00000056482.14 ENSMUST00000113996.8 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr6_+_68414401 | 0.34 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr10_-_81186137 | 0.34 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr17_-_27352876 | 0.34 |
ENSMUST00000119227.3
ENSMUST00000025045.15 |
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr6_-_87510200 | 0.33 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr12_-_115884332 | 0.33 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr12_-_114878652 | 0.32 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr3_+_96128427 | 0.32 |
ENSMUST00000090781.8
|
H2bc21
|
H2B clustered histone 21 |
| chr6_-_52181393 | 0.30 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr8_+_23464860 | 0.30 |
ENSMUST00000110688.9
ENSMUST00000121802.9 |
Ank1
|
ankyrin 1, erythroid |
| chr3_+_159201092 | 0.29 |
ENSMUST00000106041.3
|
Depdc1a
|
DEP domain containing 1a |
| chr7_-_44646960 | 0.28 |
ENSMUST00000207443.2
ENSMUST00000207755.2 ENSMUST00000003290.12 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr6_+_67532481 | 0.27 |
ENSMUST00000103302.3
|
Igkv2-137
|
immunoglobulin kappa chain variable 2-137 |
| chr16_-_92622659 | 0.27 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr6_+_70640233 | 0.27 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr2_-_160950936 | 0.26 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr6_+_92069376 | 0.26 |
ENSMUST00000113463.8
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr9_+_59485475 | 0.26 |
ENSMUST00000118549.8
ENSMUST00000034840.10 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr10_-_81186222 | 0.26 |
ENSMUST00000020454.11
ENSMUST00000105324.9 ENSMUST00000154609.3 ENSMUST00000105323.8 |
Hmg20b
|
high mobility group 20B |
| chr1_-_91386976 | 0.25 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr11_-_118977047 | 0.25 |
ENSMUST00000026665.8
|
Cbx4
|
chromobox 4 |
| chr6_-_30693675 | 0.25 |
ENSMUST00000169422.8
ENSMUST00000115131.8 ENSMUST00000115130.3 ENSMUST00000031810.15 |
Cep41
|
centrosomal protein 41 |
| chr10_+_79762858 | 0.24 |
ENSMUST00000019708.12
ENSMUST00000105377.8 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr13_-_46118433 | 0.24 |
ENSMUST00000167708.4
ENSMUST00000091628.11 ENSMUST00000180110.9 |
Atxn1
|
ataxin 1 |
| chr4_+_3678108 | 0.24 |
ENSMUST00000041377.13
ENSMUST00000103010.4 |
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr7_+_120581719 | 0.24 |
ENSMUST00000033166.10
|
Mfsd13b
|
major facilitator superfamily domain containing 13B |
| chr14_-_20784625 | 0.24 |
ENSMUST00000223679.2
|
Ndst2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr7_+_126376353 | 0.23 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr12_-_114843941 | 0.23 |
ENSMUST00000191862.6
ENSMUST00000103513.3 |
Ighv1-36
|
immunoglobulin heavy variable 1-36 |
| chr11_-_30148230 | 0.22 |
ENSMUST00000102838.10
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr18_-_79152504 | 0.22 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr12_-_113589576 | 0.21 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr12_-_114023935 | 0.21 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr12_-_115109539 | 0.21 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr5_-_46014809 | 0.20 |
ENSMUST00000190036.7
ENSMUST00000189859.7 ENSMUST00000186633.3 ENSMUST00000016026.14 ENSMUST00000045586.13 ENSMUST00000238522.2 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr9_+_31191820 | 0.19 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f2_Pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 1.5 | 7.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.1 | 4.6 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.7 | 4.9 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.6 | 1.7 | GO:1900135 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.5 | 3.8 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.4 | 2.2 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.4 | 0.9 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.4 | 2.0 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.4 | 2.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.3 | 1.4 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.3 | 1.3 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.3 | 0.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.3 | 1.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.3 | 0.8 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.3 | 1.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.3 | 1.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 2.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 2.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.5 | GO:0061344 | regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.4 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.2 | 0.6 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.2 | 1.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 0.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 1.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 0.5 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.2 | 1.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.0 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 4.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 2.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.4 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.1 | 10.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.9 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.9 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.1 | 1.6 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 1.6 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 2.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.1 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 0.3 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.2 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.7 | GO:2000111 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.3 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 3.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 3.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 1.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.6 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.1 | GO:0036019 | endolysosome(GO:0036019) |
| 0.5 | 1.4 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.4 | 1.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.3 | 2.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 4.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 0.7 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 1.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 3.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 1.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 5.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 5.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.6 | 2.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.4 | 1.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 4.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 0.6 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 3.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 4.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.9 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 2.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 5.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 3.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 2.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 10.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.9 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 9.8 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 4.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 4.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 4.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 5.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 4.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 2.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 3.2 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |