Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
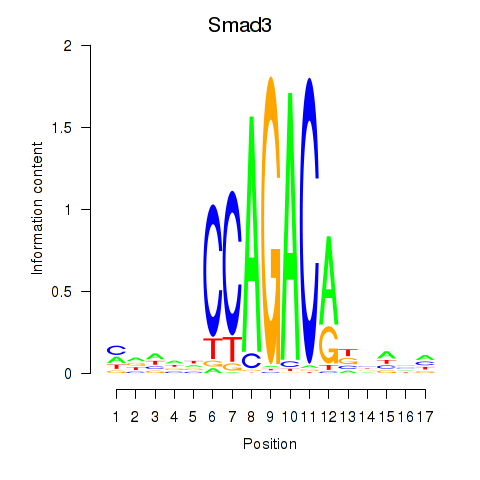
Results for Smad3
Z-value: 7.71
Transcription factors associated with Smad3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad3
|
ENSMUSG00000032402.13 | SMAD family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad3 | mm39_v1_chr9_-_63619251_63619276 | 0.86 | 6.5e-02 | Click! |
Activity profile of Smad3 motif
Sorted Z-values of Smad3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23765546 | 27.78 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr13_+_23940964 | 24.16 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr13_+_23715220 | 23.60 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr13_+_22220000 | 19.20 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr13_-_23735822 | 19.06 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr13_+_22227359 | 18.84 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr3_-_96170627 | 18.50 |
ENSMUST00000171473.3
|
H4c14
|
H4 clustered histone 14 |
| chr13_+_22017906 | 17.81 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr13_+_23718038 | 16.51 |
ENSMUST00000073261.3
|
H2ac10
|
H2A clustered histone 10 |
| chr13_-_21934675 | 16.29 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr13_+_23755551 | 13.26 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr13_-_22016364 | 12.70 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr13_+_21971631 | 11.74 |
ENSMUST00000110473.3
ENSMUST00000102982.2 |
H2bc22
|
H2B clustered histone 22 |
| chr13_-_21900313 | 10.35 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr13_-_22227114 | 9.07 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr13_-_22017677 | 8.98 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr13_-_23755374 | 8.56 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr13_-_21971388 | 6.20 |
ENSMUST00000091751.3
|
H2ac22
|
H2A clustered histone 22 |
| chr13_+_23868175 | 6.12 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr11_-_94932158 | 5.48 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr18_+_31767369 | 4.96 |
ENSMUST00000235017.2
ENSMUST00000178164.8 ENSMUST00000025109.8 |
Sap130
|
Sin3A associated protein |
| chr13_-_22225527 | 4.72 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr1_+_107456731 | 4.61 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr7_+_104506216 | 4.42 |
ENSMUST00000067695.8
|
Usp17la
|
ubiquitin specific peptidase 17-like A |
| chr13_-_23867924 | 4.31 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr13_-_42000958 | 4.12 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr7_-_100613579 | 3.85 |
ENSMUST00000060174.6
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr5_+_72360631 | 3.70 |
ENSMUST00000169617.3
|
Atp10d
|
ATPase, class V, type 10D |
| chrX_+_72454702 | 3.68 |
ENSMUST00000033740.12
|
Zfp92
|
zinc finger protein 92 |
| chr10_+_79690452 | 3.44 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr2_+_29759495 | 3.18 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr2_-_90301592 | 3.00 |
ENSMUST00000111493.8
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr15_-_103161237 | 2.99 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr1_-_21149392 | 2.80 |
ENSMUST00000037998.6
|
Tram2
|
translocating chain-associating membrane protein 2 |
| chr13_-_23015518 | 2.78 |
ENSMUST00000226294.2
ENSMUST00000226180.2 |
Vmn1r210
|
vomeronasal 1 receptor 210 |
| chr4_+_150366028 | 2.72 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr16_-_44978546 | 2.61 |
ENSMUST00000114600.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr18_+_31768126 | 2.57 |
ENSMUST00000234846.2
ENSMUST00000234772.2 |
Sap130
|
Sin3A associated protein |
| chr7_+_24920840 | 2.56 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr10_+_79690492 | 2.06 |
ENSMUST00000171599.8
ENSMUST00000095457.11 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr13_+_23191826 | 1.92 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr15_+_81820954 | 1.90 |
ENSMUST00000038757.8
ENSMUST00000230633.2 |
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr2_-_148250024 | 1.87 |
ENSMUST00000099270.5
|
Thbd
|
thrombomodulin |
| chr3_+_107008867 | 1.80 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr17_-_10538253 | 1.66 |
ENSMUST00000233828.2
ENSMUST00000233645.2 ENSMUST00000042296.9 |
Qk
|
quaking, KH domain containing RNA binding |
| chr4_+_150171822 | 1.62 |
ENSMUST00000094451.4
|
Gpr157
|
G protein-coupled receptor 157 |
| chr19_-_47907628 | 1.61 |
ENSMUST00000237029.2
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr13_+_23398297 | 1.56 |
ENSMUST00000236177.2
|
Vmn1r221
|
vomeronasal 1 receptor 221 |
| chr11_+_70323452 | 1.46 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr13_+_22268610 | 1.41 |
ENSMUST00000228243.2
ENSMUST00000226680.2 |
Vmn1r188
|
vomeronasal 1 receptor 188 |
| chr8_-_85549452 | 1.15 |
ENSMUST00000065539.6
|
Dand5
|
DAN domain family member 5, BMP antagonist |
| chr10_+_33780993 | 1.08 |
ENSMUST00000169670.8
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr6_+_136785230 | 1.06 |
ENSMUST00000074556.7
ENSMUST00000203982.2 |
H2aj
|
H2J.A histone |
| chr1_-_74076279 | 1.03 |
ENSMUST00000187281.7
|
Tns1
|
tensin 1 |
| chr5_-_53864874 | 0.99 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr2_-_132787790 | 0.95 |
ENSMUST00000038280.5
|
Fermt1
|
fermitin family member 1 |
| chr17_-_71575584 | 0.81 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr10_-_12424623 | 0.69 |
ENSMUST00000219003.2
|
Utrn
|
utrophin |
| chr13_-_42001102 | 0.68 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr3_-_27764522 | 0.62 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr14_-_50020788 | 0.60 |
ENSMUST00000118129.2
ENSMUST00000036972.14 |
Armh4
|
armadillo-like helical domain containing 4 |
| chr1_+_172168764 | 0.60 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr7_-_104019046 | 0.59 |
ENSMUST00000106831.3
|
Trim30b
|
tripartite motif-containing 30B |
| chr7_-_127187767 | 0.59 |
ENSMUST00000072155.5
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr14_+_20398230 | 0.54 |
ENSMUST00000224930.2
ENSMUST00000224110.2 ENSMUST00000225942.2 ENSMUST00000051915.7 ENSMUST00000090499.13 ENSMUST00000224721.2 ENSMUST00000090503.12 ENSMUST00000225991.2 ENSMUST00000037698.13 |
Fam149b
|
family with sequence similarity 149, member B |
| chr18_+_60659257 | 0.53 |
ENSMUST00000223984.2
ENSMUST00000025505.7 ENSMUST00000223590.2 |
Dctn4
|
dynactin 4 |
| chr8_-_13727575 | 0.48 |
ENSMUST00000117551.4
|
Rasa3
|
RAS p21 protein activator 3 |
| chr14_+_31750946 | 0.45 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr9_-_66500404 | 0.43 |
ENSMUST00000127569.8
|
Usp3
|
ubiquitin specific peptidase 3 |
| chr9_-_66500274 | 0.39 |
ENSMUST00000174387.2
|
Usp3
|
ubiquitin specific peptidase 3 |
| chr1_+_180720666 | 0.37 |
ENSMUST00000085797.6
|
Lefty2
|
left-right determination factor 2 |
| chr9_-_66500312 | 0.36 |
ENSMUST00000098613.9
|
Usp3
|
ubiquitin specific peptidase 3 |
| chr13_-_96572460 | 0.34 |
ENSMUST00000181761.2
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr3_-_79645101 | 0.28 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr4_-_129015493 | 0.28 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr5_+_34700709 | 0.26 |
ENSMUST00000101316.10
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr15_-_81581457 | 0.24 |
ENSMUST00000072910.6
|
Chadl
|
chondroadherin-like |
| chr3_+_107008343 | 0.22 |
ENSMUST00000197470.5
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr6_+_21985902 | 0.22 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr4_-_60455331 | 0.21 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr4_-_118792037 | 0.20 |
ENSMUST00000081960.5
|
Olfr1328
|
olfactory receptor 1328 |
| chr11_-_82070629 | 0.20 |
ENSMUST00000108189.9
ENSMUST00000021043.5 |
Ccl1
|
chemokine (C-C motif) ligand 1 |
| chrX_+_152615221 | 0.17 |
ENSMUST00000148708.2
ENSMUST00000123264.2 ENSMUST00000049999.9 |
Spin2c
|
spindlin family, member 2C |
| chr17_+_56935118 | 0.15 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chr7_+_26006594 | 0.15 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chrX_-_143471176 | 0.14 |
ENSMUST00000040184.4
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr5_+_105848598 | 0.14 |
ENSMUST00000120847.8
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr5_+_136067350 | 0.13 |
ENSMUST00000062606.8
|
Upk3b
|
uroplakin 3B |
| chr9_+_108867633 | 0.11 |
ENSMUST00000112059.10
ENSMUST00000026737.12 |
Shisa5
|
shisa family member 5 |
| chrX_-_7054952 | 0.11 |
ENSMUST00000004428.14
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr9_-_106533279 | 0.10 |
ENSMUST00000023959.13
ENSMUST00000201681.2 |
Grm2
|
glutamate receptor, metabotropic 2 |
| chr12_-_76842263 | 0.07 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr11_+_69047815 | 0.07 |
ENSMUST00000036424.3
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type |
| chr3_+_107538638 | 0.07 |
ENSMUST00000106703.2
|
Gm10961
|
predicted gene 10961 |
| chr11_+_87457544 | 0.07 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr2_+_3115250 | 0.06 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr10_-_120815232 | 0.06 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr7_-_30335277 | 0.06 |
ENSMUST00000108147.3
|
Etv2
|
ets variant 2 |
| chr13_-_22289994 | 0.05 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr7_-_32026670 | 0.05 |
ENSMUST00000183195.2
|
Scgb2b12
|
secretoglobin, family 2B, member 12 |
| chr7_-_34914675 | 0.05 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr11_-_102338473 | 0.05 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr7_-_32529087 | 0.04 |
ENSMUST00000182975.2
|
Scgb2b15
|
secretoglobin, family 2B, member 15 |
| chr5_-_72661546 | 0.03 |
ENSMUST00000005352.10
|
Corin
|
corin, serine peptidase |
| chr4_-_88562696 | 0.03 |
ENSMUST00000105149.3
|
Ifna13
|
interferon alpha 13 |
| chr1_-_130839178 | 0.03 |
ENSMUST00000027673.11
|
Il20
|
interleukin 20 |
| chr7_+_103065903 | 0.03 |
ENSMUST00000079348.4
|
Usp17lc
|
ubiquitin specific peptidase 17-like C |
| chr7_-_81104423 | 0.03 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr14_+_64331130 | 0.03 |
ENSMUST00000224112.2
ENSMUST00000165710.2 ENSMUST00000170709.2 |
Prss51
|
protease, serine 51 |
| chr8_+_124532781 | 0.02 |
ENSMUST00000117702.2
|
Rab4a
|
RAB4A, member RAS oncogene family |
| chr7_-_31405182 | 0.02 |
ENSMUST00000178258.3
|
Scgb2b7
|
secretoglobin, family 2B, member 7 |
| chr1_-_92408341 | 0.02 |
ENSMUST00000086837.2
|
Olfr1416
|
olfactory receptor 1416 |
| chr19_+_55886708 | 0.02 |
ENSMUST00000148666.3
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr7_-_32979763 | 0.01 |
ENSMUST00000179688.3
|
Scgb2b19
|
secretoglobin, family 2B, member 19 |
| chr7_+_123061535 | 0.01 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr7_-_102901712 | 0.01 |
ENSMUST00000073394.3
|
Usp17ld
|
ubiquitin specific peptidase 17-like D |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.7 | 2.7 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.4 | 3.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 2.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.3 | 1.0 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.3 | 1.2 | GO:1900108 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 | 2.8 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 5.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 3.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 3.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 1.0 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 1.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.6 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.7 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.3 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 1.6 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 1.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 5.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 3.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 1.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 7.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 3.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.0 | 3.8 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.5 | 1.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.4 | 5.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 1.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 3.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 2.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 2.6 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.2 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.1 | 2.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 6.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 4.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 4.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 3.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 5.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |