|
chrX_+_38754568
|
21.044
|
|
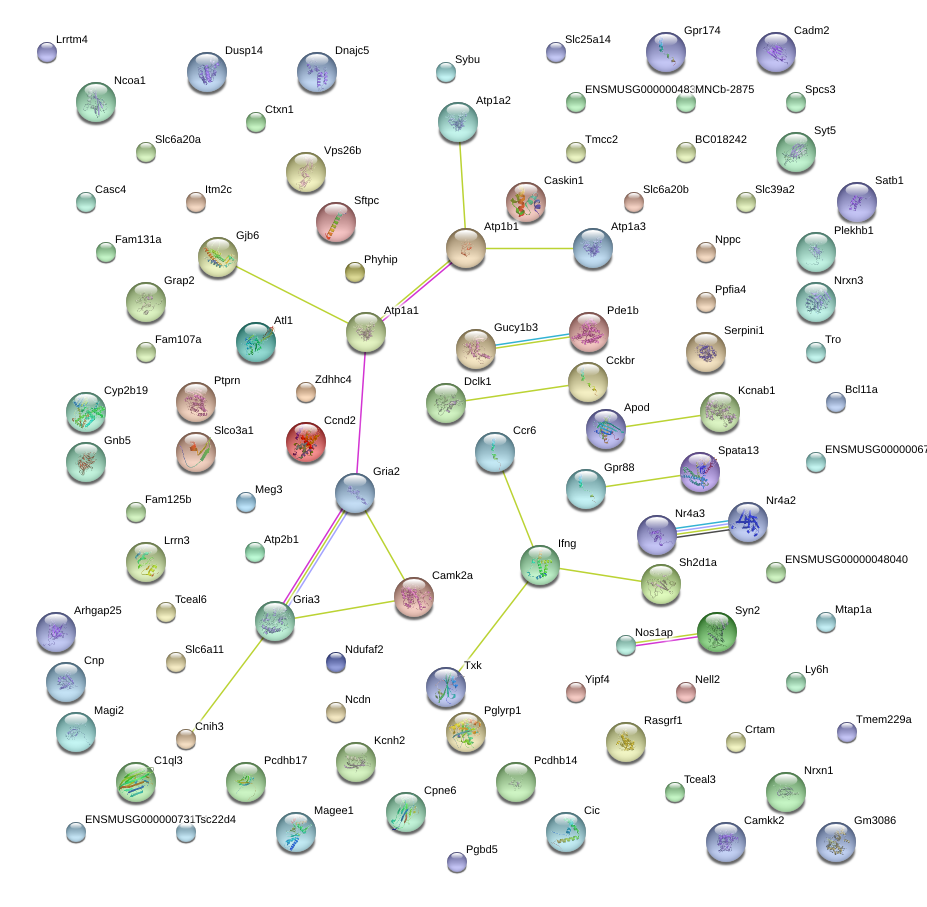
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chrX_+_38754480
|
19.442
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chrX_+_38754605
|
15.061
|
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr17_-_90436488
|
13.807
|
|
Nrxn1
|
neurexin I
|
|
chrX_+_38754658
|
12.976
|
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr1_-_75250520
|
11.320
|
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N
|
|
chr5_+_19413335
|
10.137
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr8_-_126957835
|
10.087
|
NM_171824
|
Pgbd5
|
piggyBac transposable element derived 5
|
|
chrX_+_133200982
|
9.595
|
|
Tceal3
|
transcription elongation factor A (SII)-like 3
|
|
chr7_-_4498168
|
9.418
|
NM_016908
|
Syt5
|
synaptotagmin V
|
|
chr1_-_135972341
|
9.149
|
|
|
|
|
chr7_-_107806887
|
8.773
|
NM_001163183
NM_001163184
NM_001163185
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr3_-_115955629
|
8.711
|
|
Gpr88
|
G-protein coupled receptor 88
|
|
chrX_+_133200913
|
8.412
|
NM_001029978
|
Tceal3
|
transcription elongation factor A (SII)-like 3
|
|
chr2_+_121134843
|
8.359
|
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr1_-_136229504
|
8.326
|
NM_001144855
|
Ppfia4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
|
|
chr15_-_44584003
|
8.287
|
NM_178765
|
Sybu
|
syntabulin (syntaxin-interacting)
|
|
chr12_+_71008808
|
8.194
|
|
Atl1
|
atlastin GTPase 1
|
|
chr6_+_114081227
|
8.056
|
NM_172890
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr4_+_48058102
|
7.955
|
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chrX_-_147091911
|
7.884
|
NM_001002272
NM_019548
|
Tro
|
trophinin
|
|
chr3_+_55265811
|
7.878
|
|
Dclk1
|
doublecortin-like kinase 1
|
|
chr12_+_110783786
|
7.557
|
|
Meg3
|
maternally expressed 3
|
|
chr11_+_69154265
|
7.519
|
|
A030009H04Rik
|
RIKEN cDNA A030009H04 gene
|
|
chr12_+_89961317
|
7.518
|
NM_001198587
|
Nrxn3
|
neurexin III
|
|
chr10_+_122701051
|
7.465
|
NM_182807
|
Fam19a2
|
family with sequence similarity 19, member A2
|
|
chr12_-_42212538
|
7.329
|
|
Lrrn3
|
leucine rich repeat protein 3, neuronal
|
|
chr15_+_103333554
|
7.325
|
NM_008800
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr1_-_166368439
|
7.286
|
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr17_+_8436671
|
7.161
|
NM_001190334
NM_001190335
NM_001190336
NM_001190337
|
Ccr6
|
chemokine (C-C motif) receptor 6
|
|
chr9_+_89804612
|
7.146
|
NM_001039655
NM_011245
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1
|
|
chr3_+_75361454
|
7.104
|
NM_009250
|
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1
|
|
chr12_-_4344470
|
7.093
|
|
Ncoa1
|
nuclear receptor coactivator 1
|
|
chr18_+_61123204
|
7.011
|
NM_009792
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr15_+_103333440
|
6.991
|
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr14_+_70857227
|
6.908
|
NM_145981
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein
|
|
chr10_+_98453513
|
6.818
|
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr2_+_181287207
|
6.793
|
|
Dnajc5
|
DnaJ (Hsp40) homolog, subfamily C, member 5
|
|
chr12_+_110783593
|
6.613
|
|
Meg3
|
maternally expressed 3
|
|
chr4_-_126427788
|
6.410
|
|
Ncdn
|
neurochondrin
|
|
chr9_-_26816373
|
6.269
|
|
Vps26b
|
vacuolar protein sorting 26 homolog B (yeast)
|
|
chr9_+_75159201
|
6.099
|
NM_010313
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr6_-_87483228
|
6.075
|
NM_001037727
|
Arhgap25
|
Rho GTPase activating protein 25
|
|
chrX_+_45976982
|
6.056
|
NM_001166450
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14
|
|
chr6_+_115189004
|
5.969
|
|
Syn2
|
synapsin II
|
|
chr14_+_56130147
|
5.939
|
NM_009947
|
Cpne6
|
copine VI
|
|
chr15_-_75397601
|
5.885
|
NM_011837
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr9_-_26815760
|
5.840
|
|
Vps26b
|
vacuolar protein sorting 26 homolog B (yeast)
|
|
chr8_-_4259126
|
5.785
|
NM_183315
|
Ctxn1
|
cortexin 1
|
|
chr9_+_21742717
|
5.766
|
NM_144935
|
BC018242
|
cDNA sequence BC018242
|
|
chr14_-_70923865
|
5.723
|
NM_011359
|
Sftpc
|
surfactant associated protein C
|
|
chr1_-_134287712
|
5.578
|
NM_178874
|
Tmcc2
|
transmembrane and coiled-coil domains 2
|
|
chr3_+_55265654
|
5.566
|
NM_001111051
NM_001111052
NM_001195539
NM_001195540
|
Dclk1
|
doublecortin-like kinase 1
|
|
chrX_+_39855629
|
5.536
|
NM_011364
|
Sh2d1a
|
SH2 domain protein 1A
|
|
chr6_+_79969636
|
5.519
|
NM_178731
|
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr6_-_24905939
|
5.518
|
NM_177013
|
Tmem229a
|
transmembrane protein 229A
|
|
chr7_+_26052714
|
5.485
|
|
Cic
|
capicua homolog (Drosophila)
|
|
chr7_-_25772525
|
5.468
|
|
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chrX_+_102318792
|
5.387
|
|
Magee1
|
melanoma antigen, family E, 1
|
|
chr9_-_40812668
|
5.383
|
NM_019465
|
Crtam
|
cytotoxic and regulatory T cell molecule
|
|
chr7_+_27542160
|
5.328
|
NM_007814
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19
|
|
chr5_-_73143979
|
5.267
|
NM_001122754
|
Txk
|
TXK tyrosine kinase
|
|
chr17_+_24641034
|
5.261
|
|
Caskin1
|
CASK interacting protein 1
|
|
chr2_-_91773029
|
5.249
|
|
|
|
|
chr14_-_9142289
|
5.214
|
NM_183187
|
Fam107a
|
family with sequence similarity 107, member A
|
|
chr3_-_81878554
|
5.211
|
NM_001161796
NM_017469
|
Gucy1b3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr17_-_51965912
|
5.131
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr1_+_183282758
|
5.077
|
NM_001160211
NM_001160212
NM_028408
|
Cnih3
|
cornichon homolog 3 (Drosophila)
|
|
chr11_-_83881811
|
5.074
|
NM_019819
|
Dusp14
|
dual specificity phosphatase 14
|
|
chr6_-_98877291
|
4.978
|
|
|
|
|
chr2_-_12933305
|
4.877
|
|
C1ql3
|
C1q-like 3
|
|
chr5_-_123183537
|
4.869
|
|
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chrX_+_104479684
|
4.860
|
NM_001177782
|
Gpr174
|
G protein-coupled receptor 174
|
|
chr5_+_138187303
|
4.742
|
|
Tsc22d4
|
TSC22 domain family, member 4
|
|
chr7_+_19469999
|
4.678
|
NM_009402
|
Pglyrp1
|
peptidoglycan recognition protein 1
|
|
chr14_+_52513284
|
4.628
|
NM_001039676
|
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr17_+_8435390
|
4.607
|
NM_009835
|
Ccr6
|
chemokine (C-C motif) receptor 6
|
|
chr15_-_95358578
|
4.584
|
|
Nell2
|
NEL-like 2 (chicken)
|
|
chr1_+_87804443
|
4.505
|
|
Itm2c
|
integral membrane protein 2C
|
|
chr6_-_127075853
|
4.502
|
|
Ccnd2
|
cyclin D2
|
|
chr5_-_23835349
|
4.499
|
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr16_-_31314534
|
4.494
|
NM_007470
|
Apod
|
apolipoprotein D
|
|
chr2_+_121692687
|
4.417
|
NM_177054
NM_199038
|
Casc4
|
cancer susceptibility candidate 4
|
|
chr10_+_117878101
|
4.404
|
NM_008337
|
Ifng
|
interferon gamma
|
|
chr2_-_33743465
|
4.299
|
NM_175184
|
Fam125b
|
family with sequence similarity 125, member B
|
|
chr7_-_81429262
|
4.298
|
|
Slco3a1
|
solute carrier organic anion transporter family, member 3a1
|
|
chr1_-_172487240
|
4.283
|
NM_027528
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr17_+_74898763
|
4.213
|
|
Yipf4
|
Yip1 domain family, member 4
|
|
chr11_-_6328316
|
4.209
|
|
|
|
|
chr11_+_23978110
|
4.195
|
|
Bcl11a
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr18_+_37644674
|
4.140
|
NM_053142
|
Pcdhb17
|
protocadherin beta 17
|
|
chrX_+_45976650
|
4.123
|
NM_011398
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14
|
|
chr11_+_100437204
|
4.090
|
NM_001146318
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr7_+_112574425
|
4.070
|
|
Cckbr
|
cholecystokinin B receptor
|
|
chr14_+_61351770
|
4.028
|
|
Spata13
|
spermatogenesis associated 13
|
|
chr16_-_67621054
|
4.009
|
NM_001145977
NM_178721
|
Cadm2
|
cell adhesion molecule 2
|
|
chr16_+_20695034
|
3.942
|
NM_133778
|
Fam131a
|
family with sequence similarity 131, member A
|
|
chr15_+_80453912
|
3.904
|
NM_010815
|
Grap2
|
GRB2-related adaptor protein 2
|
|
chr2_-_56967331
|
3.893
|
NM_013613
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr3_+_64913564
|
3.871
|
NM_010597
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr14_-_57744057
|
3.865
|
NM_008128
|
Gjb6
|
gap junction protein, beta 6
|
|
chr9_-_123587923
|
3.786
|
NM_139142
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A
|
|
chr8_-_55606109
|
3.738
|
|
Spcs3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae)
|
|
chr14_+_84843366
|
3.692
|
NM_001013753
|
Pcdh17
|
protocadherin 17
|
|
chr8_+_84239098
|
3.541
|
NM_001024617
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II
|
|
chr13_-_108516475
|
3.536
|
|
Zswim6
|
zinc finger, SWIM domain containing 6
|
|
chr17_-_35838974
|
3.412
|
NM_001198831
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr17_-_57217861
|
3.384
|
NM_153551
|
Dennd1c
|
DENN/MADD domain containing 1C
|
|
chr7_-_31663995
|
3.382
|
NM_009845
|
Cd22
|
CD22 antigen
|
|
chr12_+_110806715
|
3.376
|
|
Meg3
|
maternally expressed 3
|
|
chr17_-_35838818
|
3.338
|
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr12_+_3891743
|
3.337
|
NM_153743
|
Dnmt3a
|
DNA methyltransferase 3A
|
|
chr4_-_129250806
|
3.321
|
NM_001162433
|
Lck
|
lymphocyte protein tyrosine kinase
|
|
chr4_+_102432943
|
3.306
|
NM_144906
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr18_+_45719732
|
3.279
|
NM_080465
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chrX_+_156411885
|
3.274
|
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_184246785
|
3.268
|
|
Fbxo28
|
F-box protein 28
|
|
chr14_-_66211842
|
3.262
|
NM_028811
|
Elp3
|
elongation protein 3 homolog (S. cerevisiae)
|
|
chr1_+_174271338
|
3.249
|
NM_001039484
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr6_-_116666759
|
3.230
|
NM_178768
|
Tmem72
|
transmembrane protein 72
|
|
chr8_-_124999921
|
3.224
|
NM_001033142
|
Rnf166
|
ring finger protein 166
|
|
chr14_-_55735114
|
3.217
|
NM_177049
|
Jph4
|
junctophilin 4
|
|
chr18_+_45719101
|
3.213
|
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr1_-_162281701
|
3.185
|
NM_001038621
|
Rabgap1l
|
RAB GTPase activating protein 1-like
|
|
chr3_-_94386590
|
3.134
|
NM_001082484
NM_029721
|
Snx27
|
sorting nexin family member 27
|
|
chr7_-_52622323
|
3.121
|
|
|
|
|
chr17_+_21520684
|
3.059
|
NM_172486
|
Zfp677
|
zinc finger protein 677
|
|
chr3_-_117063090
|
3.048
|
|
D3Bwg0562e
|
DNA segment, Chr 3, Brigham & Women's Genetics 0562 expressed
|
|
chr15_-_95358306
|
3.030
|
|
Nell2
|
NEL-like 2 (chicken)
|
|
chr5_+_138187390
|
3.009
|
|
Tsc22d4
|
TSC22 domain family, member 4
|
|
chr1_-_174259356
|
3.009
|
NM_008429
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9
|
|
chr7_-_108729674
|
2.988
|
NM_019915
|
Art2a-ps
Art2b
|
ADP-ribosyltransferase 2a, pseudogene
ADP-ribosyltransferase 2b
|
|
chrX_-_98615554
|
2.952
|
NM_001177988
NM_019831
|
Zmym3
|
zinc finger, MYM-type 3
|
|
chr16_-_17406406
|
2.945
|
NM_001001983
|
Pi4ka
|
phosphatidylinositol 4-kinase, catalytic, alpha polypeptide
|
|
chr3_+_108163704
|
2.942
|
|
Sort1
|
sortilin 1
|
|
chr10_+_90038471
|
2.931
|
NM_001177397
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr11_+_23980676
|
2.929
|
NM_001159290
|
Bcl11a
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr14_-_68742863
|
2.909
|
|
Nefm
|
neurofilament, medium polypeptide
|
|
chr2_-_57034164
|
2.900
|
|
BB557941
|
expressed sequence BB557941
|
|
chr2_+_164648185
|
2.888
|
NM_028028
|
Zswim1
|
zinc finger, SWIM domain containing 1
|
|
chrX_+_160346948
|
2.880
|
NM_026887
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr6_+_39523866
|
2.875
|
NM_178873
|
Adck2
|
aarF domain containing kinase 2
|
|
chr6_-_60778740
|
2.844
|
NM_001042451
|
Snca
|
synuclein, alpha
|
|
chr17_+_37210011
|
2.830
|
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr4_-_153531656
|
2.826
|
|
|
|
|
chr3_-_32884000
|
2.811
|
NM_001163517
|
Pex5l
|
peroxisomal biogenesis factor 5-like
|
|
chr3_-_75360749
|
2.802
|
|
Pdcd10
|
programmed cell death 10
|
|
chr7_+_129815257
|
2.796
|
NM_019430
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr7_-_147268150
|
2.794
|
NM_001190386
|
Caly
|
calcyon neuron-specific vesicular protein
|
|
chr16_-_48772068
|
2.791
|
NM_198297
|
Trat1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr2_+_143371807
|
2.774
|
NM_008792
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr16_-_94509462
|
2.770
|
NM_139145
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase)
|
|
chr17_-_29374656
|
2.761
|
NM_153166
|
Cpne5
|
copine V
|
|
chr2_-_129525512
|
2.750
|
|
Pdyn
|
prodynorphin
|
|
chr4_+_5726856
|
2.747
|
|
Fam110b
|
family with sequence similarity 110, member B
|
|
chr7_-_90803895
|
2.744
|
|
Il16
|
interleukin 16
|
|
chr11_+_78139476
|
2.707
|
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr11_-_106163120
|
2.693
|
NM_008117
|
Gh
|
growth hormone
|
|
chr18_+_45719464
|
2.671
|
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr1_+_37426061
|
2.668
|
NM_172971
|
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I
|
|
chr16_-_28445276
|
2.667
|
NM_183064
|
Fgf12
|
fibroblast growth factor 12
|
|
chr7_+_97278722
|
2.648
|
NM_146194
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr4_+_126800830
|
2.639
|
|
Zmym6
|
zinc finger, MYM-type 6
|
|
chr16_-_19983049
|
2.630
|
NM_183390
|
Klhl6
|
kelch-like 6 (Drosophila)
|
|
chr9_-_49311823
|
2.561
|
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr5_-_124028994
|
2.537
|
|
Clip1
|
CAP-GLY domain containing linker protein 1
|
|
chr4_+_147374850
|
2.531
|
NM_008725
|
Nppa
|
natriuretic peptide type A
|
|
chr2_+_164648495
|
2.530
|
|
Zswim1
|
zinc finger, SWIM domain containing 1
|
|
chr2_+_119000244
|
2.523
|
NM_001039223
|
Gm14137
|
predicted gene 14137
|
|
chr18_+_82644510
|
2.522
|
NM_001025245
NM_010777
|
Mbp
|
myelin basic protein
|
|
chr4_+_102432879
|
2.509
|
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr1_+_89652645
|
2.509
|
NM_029846
|
Atg16l1
|
autophagy-related 16-like 1 (yeast)
|
|
chr12_+_30219769
|
2.477
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr2_+_124962427
|
2.475
|
NM_001162934
|
Ctxn2
|
cortexin 2
|
|
chr9_-_21822333
|
2.473
|
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr17_-_36408313
|
2.469
|
NM_178281
|
Trim39
|
tripartite motif-containing 39
|
|
chr7_-_52494097
|
2.418
|
NM_007645
|
Cd37
|
CD37 antigen
|
|
chr12_+_101447480
|
2.418
|
|
Calm1
|
calmodulin 1
|
|
chr6_+_48597232
|
2.399
|
NM_001077410
NM_212486
|
Gimap8
|
GTPase, IMAP family member 8
|
|
chr10_+_4978808
|
2.392
|
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr19_-_29887442
|
2.388
|
NM_177721
|
Ranbp6
|
RAN binding protein 6
|
|
chrX_+_140127827
|
2.378
|
NM_001195046
NM_001195047
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr2_+_172805342
|
2.374
|
NM_001083959
NM_001083960
NM_012046
|
Spo11
|
sporulation protein, meiosis-specific, SPO11 homolog (S. cerevisiae)
|
|
chr6_+_7505070
|
2.349
|
NM_009311
|
Tac1
|
tachykinin 1
|
|
chr19_-_7289788
|
2.346
|
|
|
|
|
chr1_+_6792987
|
2.339
|
|
St18
|
suppression of tumorigenicity 18
|
|
chr4_+_42963222
|
2.333
|
|
Dnajb5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr18_+_37644390
|
2.301
|
|
Pcdhb17
|
protocadherin beta 17
|
|
chr10_+_72119562
|
2.294
|
|
Zwint
|
ZW10 interactor
|
|
chr4_+_9196439
|
2.289
|
NM_028940
|
Clvs1
|
clavesin 1
|
|
chr2_-_25325248
|
2.287
|
NM_008963
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr6_-_36761360
|
2.263
|
NM_008973
|
Ptn
|
pleiotrophin
|
|
chr13_+_33175971
|
2.249
|
NM_173052
|
Serpinb1b
|
serine (or cysteine) peptidase inhibitor, clade B, member 1b
|
|
chrX_+_45872593
|
2.234
|
NM_011228
|
Rab33a
|
RAB33A, member of RAS oncogene family
|
|
chr10_+_115614313
|
2.233
|
NM_001161839
NM_001161838
NM_001161840
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R
|
|
chr16_+_93762803
|
2.225
|
|
Dopey2
|
dopey family member 2
|
|
chr16_-_5008915
|
2.218
|
|
|
|
|
chr7_-_85722668
|
2.213
|
NM_008746
NM_182809
|
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr2_-_12932442
|
2.205
|
NM_153155
|
C1ql3
|
C1q-like 3
|
|
chr17_-_24306262
|
2.201
|
NM_009729
|
Atp6v0c-ps2
Atp6v0c
|
ATPase, H+ transporting, lysosomal V0 subunit C, pseudogene 2
ATPase, H+ transporting, lysosomal V0 subunit C
|