ISMARA results for GNF SymAtlas, mouse, averaged across 2 replicates (Lattin, 2008)
ISMARA - Integrated System for Motif Actitivity Response Analysis is a free online tool that recognizes most important transcription factors that are changing their activity in a set of samples.
Averaging sample configuration
| Sample | Condition | Rep1_0CellLine_3T3-L1 | 0CellLine_3T3-L1 | Rep1_0CellLine_Baf3 | 0CellLine_Baf3 | Rep1_0CellLine_C2C12 | 0CellLine_C2C12 | Rep1_0CellLine_C3H_10T1_2 | 0CellLine_C3H_10T1_2 | Rep1_0CellLine_ESC_Bruce4_p13 | 0CellLine_ESC_Bruce4_p13 | Rep1_0CellLine_ESC_V26_2_p16 | 0CellLine_ESC_V26_2_p16 | Rep1_0CellLIne_mIMCD-3 | 0CellLIne_mIMCD-3 | Rep1_0CellLine_min6 | 0CellLine_min6 | Rep1_0CellLine_neuro2a | 0CellLine_neuro2a | Rep1_0CellLine_nih_3T3 | 0CellLine_nih_3T3 | Rep1_0CellLine_RAW_264_7 | 0CellLine_RAW_264_7 | Rep1_adipose_brown | adipose_brown | Rep1_adipose_white | adipose_white | Rep1_adrenal_gland | adrenal_gland | Rep1_amygdala | amygdala | Rep1_B-cells_marginal_zone | B-cells_marginal_zone | Rep1_bladder | bladder | Rep1_bone | bone | Rep1_bone_marrow | bone_marrow | Rep1_cerebellum | cerebellum | Rep1_cerebral_cortex | cerebral_cortex | Rep1_cerebral_cortex_prefrontal | cerebral_cortex_prefrontal | Rep1_ciliary_bodies | ciliary_bodies | Rep1_common_myeloid_progenitor | common_myeloid_progenitor | Rep1_cornea | cornea | Rep1_dendritic_cells_lymphoid_CD8a+ | dendritic_cells_lymphoid_CD8a+ | Rep1_dendritic_cells_myeloid_CD8a- | dendritic_cells_myeloid_CD8a- | Rep1_dendritic_plasmacytoid_B220+ | dendritic_plasmacytoid_B220+ | Rep1_dorsal_root_ganglia | dorsal_root_ganglia | Rep1_dorsal_striatum | dorsal_striatum | Rep1_epidermis | epidermis | Rep1_eyecup | eyecup | Rep1_follicular_B-cells | follicular_B-cells | Rep1_granulocytes_mac1+gr1+ | granulocytes_mac1+gr1+ | Rep1_granulo_mono_progenitor | granulo_mono_progenitor | Rep1_heart | heart | Rep1_hippocampus | hippocampus | Rep1_hypothalamus | hypothalamus | Rep1_intestine_large | intestine_large | Rep1_intestine_small | intestine_small | Rep1_iris | iris | Rep1_kidney | kidney | Rep1_lacrimal_gland | lacrimal_gland | Rep1_lens | lens | Rep1_liver | liver | Rep1_lung | lung | Rep1_lymph_nodes | lymph_nodes | Rep1_macrophage_bone_marrow_0hr | macrophage_bone_marrow_0hr | Rep1_macrophage_bone_marrow_24h_LPS | macrophage_bone_marrow_24h_LPS | Rep1_macrophage_bone_marrow_2hr_LPS | macrophage_bone_marrow_2hr_LPS | Rep1_macrophage_bone_marrow_6hr_LPS | macrophage_bone_marrow_6hr_LPS | Rep1_macrophage_peri_LPS_thio_0hrs | macrophage_peri_LPS_thio_0hrs | Rep1_macrophage_peri_LPS_thio_1hrs | macrophage_peri_LPS_thio_1hrs | Rep1_macrophage_peri_LPS_thio_7hrs | macrophage_peri_LPS_thio_7hrs | Rep1_mammary_gland__lact | mammary_gland__lact | Rep1_mammary_gland_non-lactating | mammary_gland_non-lactating | Rep1_mast_cells | mast_cells | Rep1_mast_cells_IgE+antigen_1hr | mast_cells_IgE+antigen_1hr | Rep1_mast_cells_IgE+antigen_6hr | mast_cells_IgE+antigen_6hr | Rep1_mast_cells_IgE | mast_cells_IgE | Rep1_mega_erythrocyte_progenitor | mega_erythrocyte_progenitor | Rep1_microglia | microglia | Rep1_NK_cells | NK_cells | Rep1_nucleus_accumbens | nucleus_accumbens | Rep1_olfactory_bulb | olfactory_bulb | Rep1_osteoblast_day14 | osteoblast_day14 | Rep1_osteoblast_day21 | osteoblast_day21 | Rep1_osteoblast_day5 | osteoblast_day5 | Rep1_osteoclasts | osteoclasts | Rep1_ovary | ovary | Rep1_pancreas | pancreas | Rep1_pituitary | pituitary | Rep1_placenta | placenta | Rep1_prostate | prostate | Rep1_retina | retina | Rep1_retinal_pigment_epithelium | retinal_pigment_epithelium | Rep1_salivary_gland | salivary_gland | Rep1_skeletal_muscle | skeletal_muscle | Rep1_spinal_cord | spinal_cord | Rep1_spleen | spleen | Rep1_stem_cells__HSC | stem_cells__HSC | Rep1_stomach | stomach | Rep1_T-cells_CD4+ | T-cells_CD4+ | Rep1_T-cells_CD8+ | T-cells_CD8+ | Rep1_T-cells_foxP3+ | T-cells_foxP3+ | Rep1_testis | testis | Rep1_thymocyte_DP_CD4+CD8+ | thymocyte_DP_CD4+CD8+ | Rep1_thymocyte_SP_CD4+ | thymocyte_SP_CD4+ | Rep1_thymocyte_SP_CD8+ | thymocyte_SP_CD8+ | Rep1_umbilical_cord | umbilical_cord | Rep1_uterus | uterus | Rep2_0CellLine_3T3-L1 | 0CellLine_3T3-L1 | Rep2_0CellLine_Baf3 | 0CellLine_Baf3 | Rep2_0CellLine_C2C12 | 0CellLine_C2C12 | Rep2_0CellLine_C3H_10T1_2 | 0CellLine_C3H_10T1_2 | Rep2_0CellLine_ESC_Bruce4_p13 | 0CellLine_ESC_Bruce4_p13 | Rep2_0CellLine_ESC_V26_2_p16 | 0CellLine_ESC_V26_2_p16 | Rep2_0CellLine_mIMCD-3 | 0CellLIne_mIMCD-3 | Rep2_0CellLine_min6 | 0CellLine_min6 | Rep2_0CellLine_neuro2a | 0CellLine_neuro2a | Rep2_0CellLine_nih_3T3 | 0CellLine_nih_3T3 | Rep2_0CellLine_RAW_264_7 | 0CellLine_RAW_264_7 | Rep2_adipose_brown | adipose_brown | Rep2_adipose_white | adipose_white | Rep2_adrenal_gland | adrenal_gland | Rep2_amygdala | amygdala | Rep2_B-cells_marginal_zone | B-cells_marginal_zone | Rep2_bladder | bladder | Rep2_bone | bone | Rep2_bone_marrow | bone_marrow | Rep2_cerebellum | cerebellum | Rep2_cerebral_cortex | cerebral_cortex | Rep2_cerebral_cortex_prefrontal | cerebral_cortex_prefrontal | Rep2_ciliary_bodies | ciliary_bodies | Rep2_common_myeloid_progenitor | common_myeloid_progenitor | Rep2_cornea | cornea | Rep2_dendritic_cells_lymphoid_CD8a+ | dendritic_cells_lymphoid_CD8a+ | Rep2_dendritic_cells_myeloid_CD8a- | dendritic_cells_myeloid_CD8a- | Rep2_dendritic_plasmacytoid_B220+ | dendritic_plasmacytoid_B220+ | Rep2_dorsal_root_ganglia | dorsal_root_ganglia | Rep2_dorsal_striatum | dorsal_striatum | Rep2_epidermis | epidermis | Rep2_eyecup | eyecup | Rep2_follicular_B-cells | follicular_B-cells | Rep2_granulocytes_mac1+gr1+ | granulocytes_mac1+gr1+ | Rep2_granulo_mono_progenitor | granulo_mono_progenitor | Rep2_heart | heart | Rep2_hippocampus | hippocampus | Rep2_hypothalamus | hypothalamus | Rep2_intestine_large | intestine_large | Rep2_intestine_small | intestine_small | Rep2_iris | iris | Rep2_kidney | kidney | Rep2_lacrimal_gland | lacrimal_gland | Rep2_lens | lens | Rep2_liver | liver | Rep2_lung | lung | Rep2_lymph_nodes | lymph_nodes | Rep2_macrophage_bone_marrow_0hr | macrophage_bone_marrow_0hr | Rep2_macrophage_bone_marrow_24h_LPS | macrophage_bone_marrow_24h_LPS | Rep2_macrophage_bone_marrow_2hr_LPS | macrophage_bone_marrow_2hr_LPS | Rep2_macrophage_bone_marrow_6hr_LPS | macrophage_bone_marrow_6hr_LPS | Rep2_macrophage_peri_LPS_thio_0hrs | macrophage_peri_LPS_thio_0hrs | Rep2_macrophage_peri_LPS_thio_1hrs | macrophage_peri_LPS_thio_1hrs | Rep2_macrophage_peri_LPS_thio_7hrs | macrophage_peri_LPS_thio_7hrs | Rep2_mammary_gland_0lact | mammary_gland__lact | Rep2_mammary_gland_non-lactating | mammary_gland_non-lactating | Rep2_mast_cells | mast_cells | Rep2_mast_cells_IgE+antigen_1hr | mast_cells_IgE+antigen_1hr | Rep2_mast_cells_IgE+antigen_6hr | mast_cells_IgE+antigen_6hr | Rep2_mast_cells_IgE | mast_cells_IgE | Rep2_mega_erythrocyte_progenitor | mega_erythrocyte_progenitor | Rep2_microglia | microglia | Rep2_NK_cells | NK_cells | Rep2_nucleus_accumbens | nucleus_accumbens | Rep2_olfactory_bulb | olfactory_bulb | Rep2_osteoblast_day14 | osteoblast_day14 | Rep2_osteoblast_day21 | osteoblast_day21 | Rep2_osteoblast_day5 | osteoblast_day5 | Rep2_osteoclasts | osteoclasts | Rep2_ovary | ovary | Rep2_pancreas | pancreas | Rep2_pituitary | pituitary | Rep2_placenta | placenta | Rep2_prostate | prostate | Rep2_retina | retina | Rep2_retinal_pigment_epithelium | retinal_pigment_epithelium | Rep2_salivary_gland | salivary_gland | Rep2_skeletal_muscle | skeletal_muscle | Rep2_spinal_cord | spinal_cord | Rep2_spleen | spleen | Rep2_stem_cells_0HSC | stem_cells__HSC | Rep2_stomach | stomach | Rep2_T-cells_CD4+ | T-cells_CD4+ | Rep2_T-cells_CD8+ | T-cells_CD8+ | Rep2_T-cells_foxP3+ | T-cells_foxP3+ | Rep2_testis | testis | Rep2_thymocyte_DP_CD4+CD8+ | thymocyte_DP_CD4+CD8+ | Rep2_thymocyte_SP_CD4+ | thymocyte_SP_CD4+ | Rep2_thymocyte_SP_CD8+ | thymocyte_SP_CD8+ | Rep2_umbilical_cord | umbilical_cord | Rep2_uterus | uterus |
|---|
All motifs sorted by activity significance
| Motif name | Z-value | Associated genes | Profile | Logo |
|---|---|---|---|---|
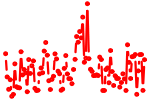
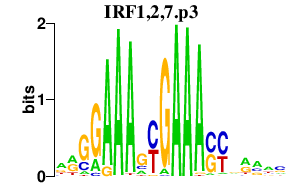
| IRF1,2,7.p3 | 9.831 |
Irf1
(Irf-1)
Irf2 (Irf-2) Irf7 |
|
|
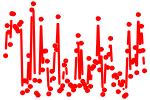
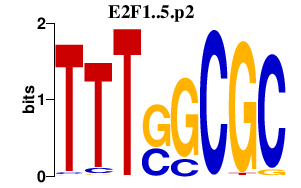
| E2F1..5.p2 | 8.685 |
E2f4
E2f5 (E2F-5) E2f2 E2f1 (E2F-1) E2f3 (E2F3b, E2f3a) |
|
|
| RFX1..5_RFXANK_RFXAP.p2 | 7.803 |
Rfx1
Rfxap Rfx5 Rfxank (Tvl1) Rfx4 Rfx3 (MRFX3) Rfx2 |
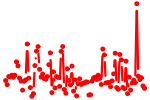
|
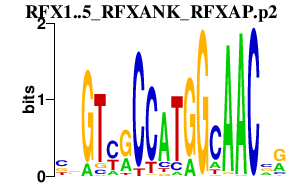
|
| REST.p3 | 7.570 |
Rest
(NRSF)
|
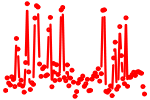
|
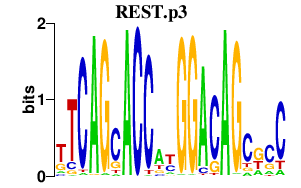
|
| SPIB.p2 | 7.147 |
Spib
(Spi-B)
|

|
|
| NRF1.p2 | 6.605 |
Nrf1
(D6Ertd415e)
|

|
|
| HNF1A.p2 | 6.257 |
Hnf1a
(hepatocyte nuclear factor 1, HNF1, Hnf-1, Hnf1alpha, HNF1-alpha, LFB1, Tcf1, HNF1[a])
|
|
|
| GATA1..3.p2 | 6.107 |
Gata1
(Gata-1, Gf-1)
Gata2 (Gata-2) Gata3 (Gata-3) |

|

|
| TEAD1.p2 | 6.042 |
Tead1
(Gtrgeo5, TEAD-1, TEF-1, mTEF-1, Tcf13)
Tead2 (Etdf, TEF-4, TEAD-2) Tead3 (TEAD-3, Tcf13r2, TEF-5, DTEF-1, ETFR-1) Tead4 (ETFR-2a, TEF-3, Tef3, TEAD-4, Etfr2, Tefr, Tcf13r1, Tefr1a) |

|
|
| SPI1.p2 | 6.014 |
Sfpi1
(Sfpi-1, PU.1, Spi-1, Tcfpu1, Dis-1, Tfpu.1)
|

|
|
| NFKB1_REL_RELA.p2 | 5.926 |
Rela
(p65 NF kappaB, p65)
Rel (c-Rel) Nfkb1 (nuclear factor kappaB p50, NF-kappaB p50, p50, NF-kappaB, p50/p105, p50 subunit of NF kappaB, NF kappaB1) |
|
|
| MEF2{A,B,C,D}.p2 | 5.907 |
Mef2a
Mef2c Mef2d Mef2b |
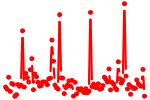
|
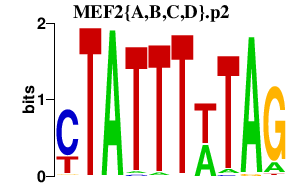
|
| HOX{A6,A7,B6,B7}.p2 | 5.556 |
Hoxb6
(Hox-2.2)
Hoxa7 (Hox-1.1) Hoxb7 (Hox-2.3) Hoxa6 (Hox-1.2) |
|

|
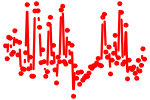
| ELK1,4_GABP{A,B1}.p3 | 5.498 |
Gabpa
(GABPalpha)
Gabpb1 (NRF2B2, E4TF1-47, BABPB2, E4TF1, NRF2B1, E4Tf1B, GABPB1-1, GABPB1-2, E4TF1-53) Elk4 (Sap1) Elk1 (Elk-1) |

|
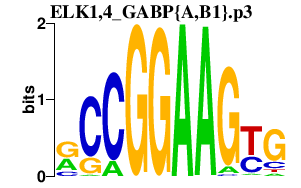
|
| YY1.p2 | 5.429 |
Yy1
(Yin Yang 1, delta transcription factor, UCRBP transcription factor, NF-E1)
|

|
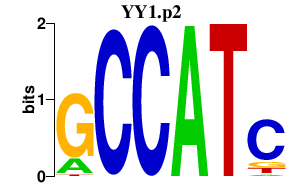
|
| MYB.p2 | 5.407 |
Myb
(c-myb)
|

|
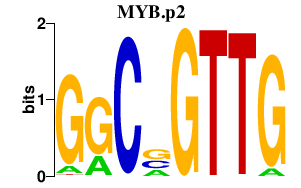
|
| ZNF143.p2 | 5.137 |
Zfp143
(KRAB14, Zfp80-rs1, Zfp79, Staf, D7Ertd805e, pHZ-1)
|

|
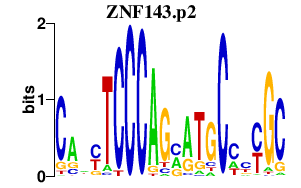
|
| TBP.p2 | 5.021 |
Tbp
(Gtf2d)
|

|
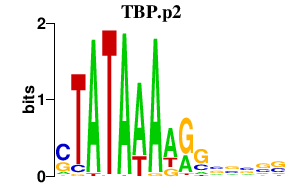
|
| XBP1.p3 | 5.003 |
Xbp1
(D11Ertd39e, TREB5, TREB-5, XBP-1)
|

|
|
| TLX1..3_NFIC{dimer}.p2 | 4.943 |
Tlx1
(Hox-11, Hox11)
Tlx3 (Rnx, Tlx1l2, Hox11l2) |

|
|
| ELF1,2,4.p2 | 4.942 |
Elf2
(NERF-2)
Elf1 (p70, Elf-1, mElf-1) Elf4 (myeloid elf-1 like factor, MEF) |

|
|
| HNF4A_NR2F1,2.p2 | 4.812 |
Nr2f1
(COUP-TF1, Tcfcoup1, Erbal3, COUP-TFI)
Nr2f2 (Tcfcoup2, COUP-TFII, ARP-1, EAR3, COUP-TF2, Aporp1) Hnf4a (Tcf4, Hnf4, Tcf14, Nuclear receptor 2A1, Nr2a1, HNF4 alpha, HNF-4, MODY1) |
|
|
| NR5A1,2.p2 | 4.681 |
Nr5a1
(Ad4BP, ELP, adrenal 4-binding protein, SF1, Ftz-F1, Ftzf1, SF-1, steroidogenic factor 1)
Nr5a2 (D1Ertd308e, Ftf, LRH-1) |

|
|
| SRF.p3 | 4.631 |
Srf
|
|
|
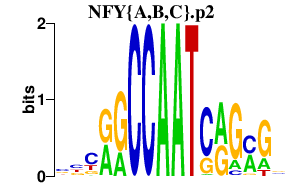
| NFY{A,B,C}.p2 | 4.605 |
Nfyc
Nfyb (Cbf-A) Nfya (Sez10, Cbf-b) |
|
|
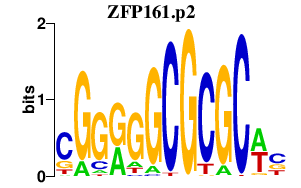
| ZFP161.p2 | 4.481 |
Zfp161
(ZF5)
|

|
|
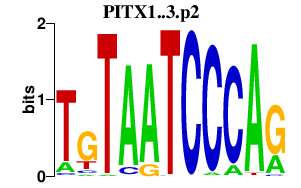
| PITX1..3.p2 | 4.439 |
Pitx2
(solurshin, Munc30, Brx1, Brx1a, Pitx2a, Pitx2b, Brx1b, Pitx2c, Otlx2, Rieg, Ptx2)
Pitx3 (Ptx3) Pitx1 (P-OTX, Potx, Bft, Ptx1) |

|
|
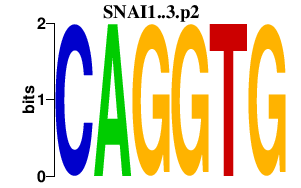
| SNAI1..3.p2 | 4.322 |
Snai2
(Snail2, Slugh, Slug)
Snai1 (Sna, Snail1, Snail, Sna1) Snai3 (Smuc, Snail-related gene from muscle cells, Zfp293) |
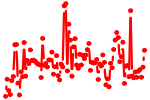
|
|
| MAFB.p2 | 4.297 |
Mafb
(Krml, Kreisler, Krml1)
|

|
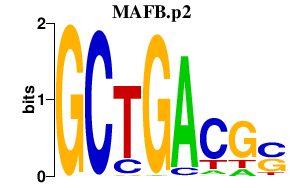
|
| GATA6.p2 | 4.288 |
Gata6
|
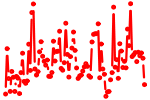
|
|
| RUNX1..3.p2 | 4.220 |
Runx1
(AML1, runt domain, alpha subunit 2, Pebp2a2, Cbfa2)
Runx3 (Cbfa3, AML2) Runx2 (SL3-3 enhancer factor 1, AML3, polyomavirus enhancer binding factor 2 (PEBP2), PEBP2 alpha A, PEBP2aA, Cbfa1, Pebpa2a, Osf2) |

|
|
| TFCP2.p2 | 4.107 |
Tcfcp2
(UBP-1, CP-2, CP2, LBP-1d, LBP1, LBP-1c, LSF)
|
|
|
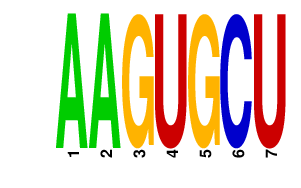
| UGUGCUU | 4.097 | mmu-miR-218 |
|

|
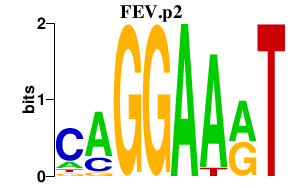
| FEV.p2 | 4.035 |
Fev
(Pet1, mPet-1)
|

|
|
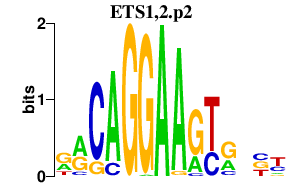
| ETS1,2.p2 | 3.992 |
Ets2
(Ets-2)
Ets1 (Tpl1, p51Ets-1, p42Ets-1, Ets-1) |

|
|
| ESRRA.p2 | 3.971 |
Esrra
(ERRalpha, Estrra, Err1, Nr3b1)
|
|

|
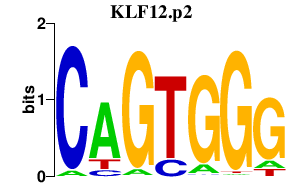
| KLF12.p2 | 3.913 |
Klf12
(AP-2rep)
|

|
|
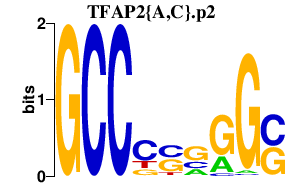
| TFAP2{A,C}.p2 | 3.879 |
Tcfap2c
(AP2gamma, Ap-2.2, Stra2)
Tcfap2a (AP-2 alpha, AP2alpha, Ap2tf, Ap-2 (a), Ap2) |
|
|
| KLF4.p3 | 3.878 |
Klf4
(EZF, Zie, Gklf)
|
|
|
| MTF1.p2 | 3.785 |
Mtf1
(MTF-1, metal response element-binding transcription factor 1, metalloregulatory transcription factor, Thyls)
|

|
|
| TFDP1.p2 | 3.696 |
Tfdp1
(Dp1, Drtf1)
|

|

|
| AAGGCAC | 3.696 | mmu-miR-124 |

|

|
| HBP1_HMGB_SSRP1_UBTF.p2 | 3.631 |
Hmgb2
(Hmg2, HMG-2)
Hmgb3 (Hmg2a, Hmg4) Ssrp1 (Hmg1-rs1, Hmgox, Hmgi-rs3, T160) Ubtf (UBF1, UBF, Tcfubf) Hbp1 (C86454) |

|
|
| ZEB1.p2 | 3.517 |
Zeb1
(Zfx1a, Zfhx1a, MEB1, AREB6, [delta]EF1, ZEB, Tcf8, Zfhep, Tcf18, Nil2)
|

|
|
| FOS_FOS{B,L1}_JUN{B,D}.p2 | 3.504 |
Fos
(cFos, c-fos, D12Rfj1)
Junb Fosl1 (fra-1, Fra1) Jund Fosb |

|

|
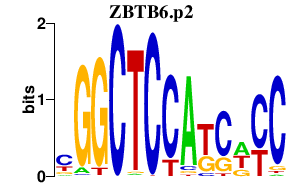
| ZBTB6.p2 | 3.363 |
Zbtb6
(Zfp482)
|

|
|
| AGCACCA | 3.350 | mmu-miR-29a mmu-miR-29b mmu-miR-29c |
|

|
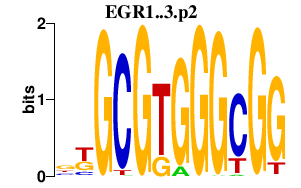
| EGR1..3.p2 | 3.316 |
Egr3
(Pilot)
Egr1 (NGF1-A, NGFIA, Zenk, Egr-1, Krox-1, ETR103, TIS8, Krox-24, Zif268, Zfp-6, NGFI-A, Krox24) Egr2 (Egr-2, Krox-20, Zfp-25, NGF1-B, Krox20) |

|
|
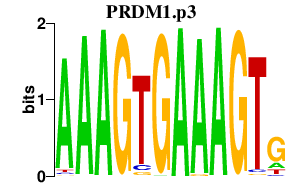
| PRDM1.p3 | 3.311 |
Prdm1
(PRDI-BF1, Blimp-1, Blimp1)
|

|
|
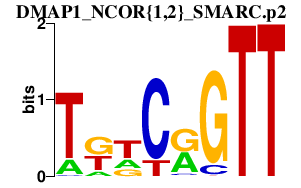
| DMAP1_NCOR{1,2}_SMARC.p2 | 3.310 |
Smarca1
(Snf2l)
Smarcc2 Smarca5 (Snf2h, MommeD4) Ncor1 (N-CoR, Rxrip13) Ncor2 (SMRTe, SMRT) Dmap1 |

|
|
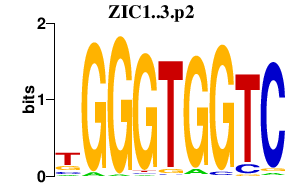
| ZIC1..3.p2 | 3.281 |
Zic1
(odd-paired homolog)
Zic2 (odd-paired homolog, Ku, GENA 29) Zic3 |

|
|
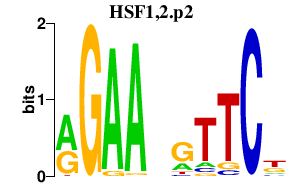
| HSF1,2.p2 | 3.255 |
Hsf1
(heat shock transcription factor 1)
Hsf2 |

|
|
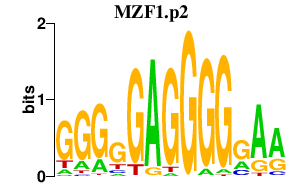
| MZF1.p2 | 3.219 |
Mzf1
(Znf42, Mzf2, Zfp121, Zfp98)
|

|
|
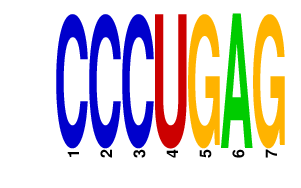
| CCCUGAG | 3.136 | mmu-miR-125a-5p mmu-miR-125b-5p mmu-miR-351 |

|
|
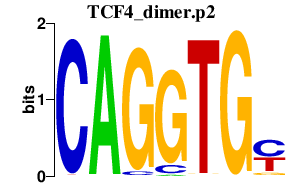
| TCF4_dimer.p2 | 3.127 |
Tcf4
(E2.2, bHLHb19, SEF-2, TFE, ME2, ITF-2b, ASP-I2, MITF-2A, SEF2-1, E2-2, ITF-2, MITF-2B)
|

|
|
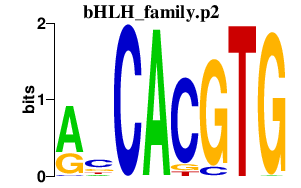
| bHLH_family.p2 | 3.085 |
Arntl
(Bmal1, MOP3, bHLHe5, Arnt3)
Mitf (bHLHe32, wh, mi) Mxi1 (Mad2, bHLHc11) Tcfe3 (Tfe-3, bHLHe33, Tfe3) Clock (bHLHe8, KAT13D) Olig2 (Olg-2, Bhlhb1, bHLHe19, RK17) Mxd4 (Mad4, bHLHc12) Hey2 (Hrt2, bHLHb32, Hesr2, Herp1, CHF1) Hey1 (bHLHb31, Hesr1, hesr-1, Herp2, HRT1) Heyl (Hey3, Hrt3, bHLHb33, Hesr3) Mlxipl (bHLHd14, Wbscr14, ChREBP, WS-bHLH) Hes6 (bHLHb41) Mxd3 (bHLHc13, Mad3) Arntl2 (MOP9, bHLHe6, BMAL2) Id1 (Idb1, D2Wsu140e, bHLHb24) Mnt (Rox, bHLHd3) Olig1 (Olg-1, Bhlhb6, bHLHe21) Npas2 (bHLHe9, MOP4) |
|
|
| GFI1.p2 | 3.062 |
Gfi1
(Pal-1, Gfi-1, Pal1)
|

|
|
| FOX{F1,F2,J1}.p2 | 3.031 |
Foxf2
(Fkh20, LUN, FREAC2)
Foxj1 (HFH-4, Hfh4, FKHL-13) Foxf1a (HFH-8, Hfh8, Freac-1, FREAC1, Foxf1) |

|
|
| ATF4.p2 | 2.990 |
Atf4
(CREB2, Atf-4, C/ATF, TAXREB67)
|

|
|
| MYOD1.p2 | 2.989 |
Myod1
(Myod-1, bHLHc1, MyoD, MYF3)
|

|
|
| TGIF1.p2 | 2.968 |
Tgif1
(Tgif)
|

|
|
| AHR_ARNT_ARNT2.p2 | 2.963 |
Arnt
(D3Ertd557e, bHLHe2, ESTM42, Hif1b)
Ahr (Ahre, dioxin receptor, bHLHe76, Ahh, In, Ah) Arnt2 (bHLHe1) |

|

|
| ATF2.p2 | 2.916 |
Atf2
(Creb2, ATF-2, mXBP, CRE-BP)
|

|
|
| ARNT_ARNT2_BHLHB2_MAX_MYC_USF1.p2 | 2.908 |
Arnt
(D3Ertd557e, bHLHe2, ESTM42, Hif1b)
Max (bHLHd7, bHLHd5, bHLHd8, bHLHd6, bHLHd4) Myc (Niard, Nird, bHLHe39, Myc2, c-myc) Usf1 (bHLHb11, upstream stimulatory factor) Bhlhe40 (eip1 (E47 interaction protein 1), Stra13, DEC1, Clast5, Bhlhb2, cytokine response gene 8, Stra14, CR8, Sharp2) Arnt2 (bHLHe1) |
|
|
| GGCAGUG | 2.904 | mmu-miR-34a mmu-miR-34b-5p mmu-miR-34c mmu-miR-449a mmu-miR-449b mmu-miR-449c |
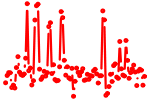
|
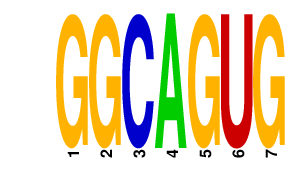
|
| NFE2L1.p2 | 2.892 |
Nfe2l1
(TCF-11, TCF11, NRF1, LCR-F1, Lcrf1)
|

|

|
| ZNF238.p2 | 2.879 |
Zfp238
(RP58)
|

|
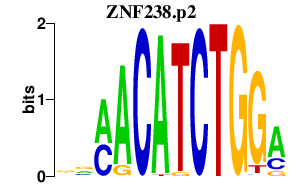
|
| FOXA2.p3 | 2.843 |
Foxa2
(Hnf3b, HNF3beta, HNF3-beta, Tcf3b, Hnf-3b, Tcf-3b)
|
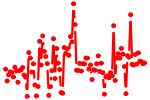
|

|
| GTF2I.p2 | 2.817 |
Gtf2i
(TFII-I, BAP-135)
|
|

|
| FOXD3.p2 | 2.816 |
Foxd3
(Hfh2, Genesis, CWH3)
|

|

|
| LMO2.p2 | 2.773 |
Lmo2
(Rbtn-2, Rbtn2, Rhom-2)
|

|
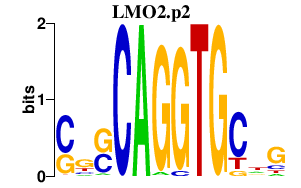
|
| MYFfamily.p2 | 2.751 |
Myod1
(Myod-1, bHLHc1, MyoD, MYF3)
Myf6 (herculin, MRF4, bHLHc4) Myf5 (bHLHc2, Myf-5) Myog (bHLHc3, myo) |

|
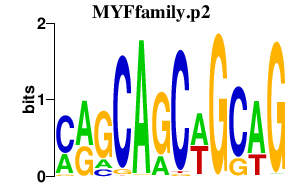
|
| FOX{D1,D2}.p2 | 2.712 |
Foxd2
(Mf2)
Foxd1 (FREAC4, BF-2, Hfh10, Hfhbf2) |
|
|
| JUN.p2 | 2.684 |
Jun
(c-jun, Junc)
Jund Junb |

|
|
| EHF.p2 | 2.670 |
Ehf
|

|
|
| BPTF.p2 | 2.663 |
Bptf
(Falz)
|

|
|
| UAUUGCU | 2.509 | mmu-miR-137 |
|

|
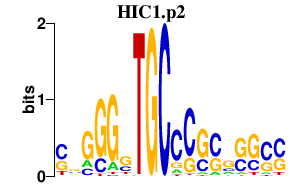
| HIC1.p2 | 2.505 |
Hic1
(HIC-1)
|

|
|
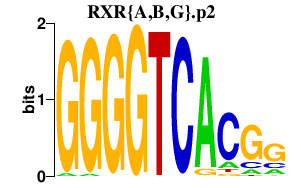
| RXR{A,B,G}.p2 | 2.504 |
Rxra
(RXR alpha 1, RXRalpha1)
Rxrb (Nr2b2, H-2RIIBP, RCoR-1, Rub) Rxrg (Nr2b3) |

|
|
| CUUUGGU | 2.481 | mmu-miR-9 |

|

|
| NFATC1..3.p2 | 2.471 |
Nfatc3
(D8Ertd281e, NFATx, NFAT4)
Nfatc2 (NFAT1-D, NFAT1, NFATp) Nfatc1 (NFATc, NFAT2, NF-ATc) |

|

|
| ATF5_CREB3.p2 | 2.463 |
Atf5
(Atf7, AFTA, Atfx, ODA-10)
Creb3 (LZIP-1, LZIP, Luman, LZIP-2) |
|
|
| MAZ.p2 | 2.451 |
Maz
(Pur-1, SAF-1, PUR1, SAF-2)
|

|
|
| HIF1A.p2 | 2.431 |
Hif1a
(MOP1, bHLHe78, HIF-1alpha, HIF1alpha)
|

|
|
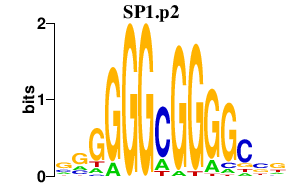
| SP1.p2 | 2.400 |
Sp1
(Sp1-1)
|
|
|
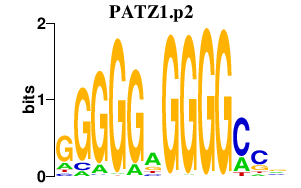
| PATZ1.p2 | 2.379 |
Patz1
(Zfp278, MAZR, POZ-AT hook-zinc finger protein)
|
|
|
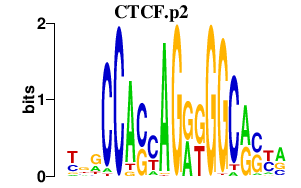
| CTCF.p2 | 2.367 |
Ctcf
|

|
|
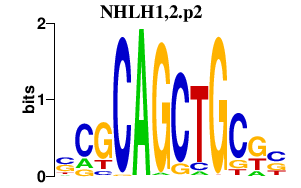
| NHLH1,2.p2 | 2.328 |
Nhlh1
(bHLHa35, Hen1, Tal2, Nscl)
Nhlh2 (bHLHa34, NSCL2, Nscl-2, Hen2) |

|
|
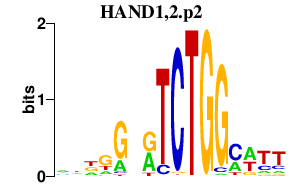
| HAND1,2.p2 | 2.309 |
Hand1
(Thing1, Hxt, eHAND, bHLHa27, Th1, Ehand1)
Hand2 (bHLHa26, Thing2, Hed, Ehand2, dHAND) |
|
|
| ATF6.p2 | 2.301 |
Atf6
(ESTM49, Atf6alpha)
|
|

|
| GUAGUGU | 2.288 | mmu-miR-142-3p |

|

|
| UUGGUCC | 2.271 | mmu-miR-133a mmu-miR-133b |

|

|
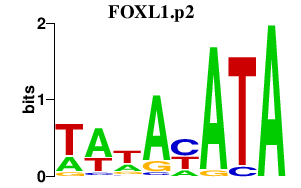
| FOXL1.p2 | 2.243 |
Foxl1
(fkh6, Fkh6, FREAC7, fkh-6)
|

|
|
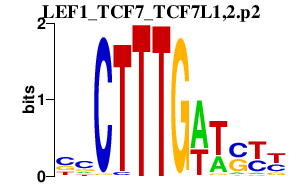
| LEF1_TCF7_TCF7L1,2.p2 | 2.234 |
Tcf7
(TCF-1, T-cell factor 1, Tcf1, T cell factor-1)
Tcf7l2 (mTcf-4E, Tcf4, mTcf-4B, TCF4E, Tcf-4, TCF4B) Lef1 (Lef-1, lymphoid enhancer factor 1) Tcf7l1 (Tcf3) |
|
|
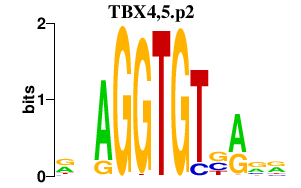
| TBX4,5.p2 | 2.232 |
Tbx5
Tbx4 |

|
|
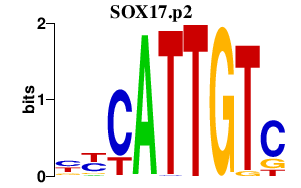
| SOX17.p2 | 2.225 |
Sox17
|

|
|
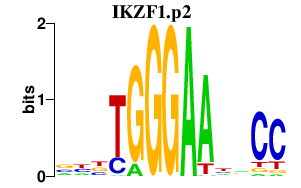
| IKZF1.p2 | 2.224 |
Ikzf1
(Ikaros, Zfpn1a1, LyF-1)
|
|
|
| NR1H4.p2 | 2.212 |
Nr1h4
(RIP14, Rxrip14, HRR1, Fxr, FXR)
|
|
|
| POU5F1.p2 | 2.203 |
Pou5f1
(Otf3-rs7, Oct3/4, Otf-4, Oct-3, Oct4, Otf3, Oct-3/4, Otf3g, Otf-3, Otf4, Oct-4)
|

|

|
| TFEB.p2 | 2.200 |
Tcfeb
(bHLHe35, TFEB)
|

|
|
| TLX2.p2 | 2.198 |
Tlx2
(Ncx1, Enx, Hox11L.1, Tlx1l1, NCX, Hox11l1)
|
|
|
| AACACUG | 2.193 | mmu-miR-141 mmu-miR-200a |

|

|
| UCCAGUU | 2.193 | mmu-miR-145 |
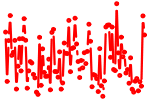
|

|
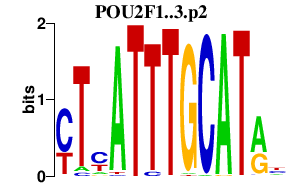
| POU2F1..3.p2 | 2.189 |
Pou2f2
(Otf-2, Oct2b, Oct2a, Oct-2, Otf2)
Pou2f1 (Oct-1C, Oct-1z, Oct1, Otf1, Oct-1A, Otf-1, oct-1, Oct-1B) Pou2f3 (Skin-1a, Skn-li, Skn-1a, Skin, Oct11, Otf11, Oct-11a, Otf-11, Epoc-1) |

|
|
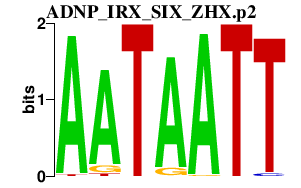
| ADNP_IRX_SIX_ZHX.p2 | 2.188 |
Six5
(MDMAHP, TrexBF, Dmahp)
Zhx1 Irx4 Adnp Zhx3 (Tix1) Zhx2 (Raf, Afr-1, Afr1) Irx5 Six2 |

|
|
| AAUACUG | 2.185 | mmu-miR-200b mmu-miR-200c mmu-miR-429 |

|

|
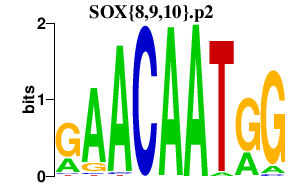
| SOX{8,9,10}.p2 | 2.175 |
Sox9
Sox8 Sox10 (Sox21) |

|
|
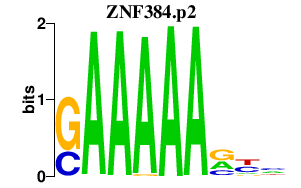
| ZNF384.p2 | 2.175 |
Zfp384
(Nmp4, Ciz)
|

|
|
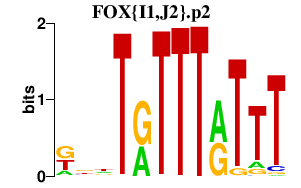
| FOX{I1,J2}.p2 | 2.172 |
Foxi1
(HFH-3, Fkh10, Hfh3)
Foxj2 (Fhx) |

|
|
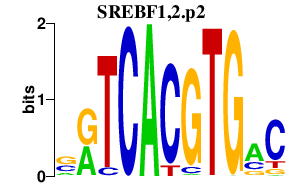
| SREBF1,2.p2 | 2.154 |
Srebf1
(SREBP-1a, SREBP1, SREBP-1c, SREBP-1, bHLHd1, ADD-1, SREBP1c)
Srebf2 (bHLHd2, SREBP-2, SREBP2gc, SREBP2) |

|
|
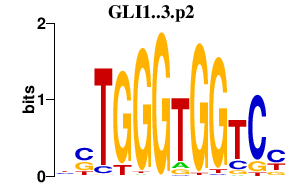
| GLI1..3.p2 | 2.137 |
Gli1
(Zfp-5, Zfp5)
Gli2 Gli3 (Bph, brachyphalangy) |

|
|
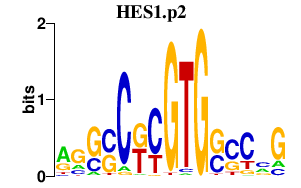
| HES1.p2 | 2.121 |
Hes1
(Hry, bHLHb39)
|

|
|
| PRRX1,2.p2 | 2.107 |
Prrx2
(S8, Prx2)
Prrx1 (Pmx1, Prx1, mHox, K-2) |

|

|
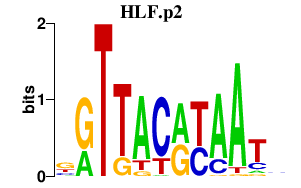
| HLF.p2 | 2.100 |
Hlf
|
|
|
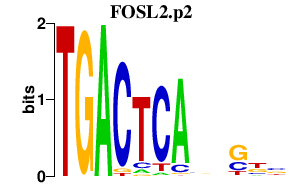
| FOSL2.p2 | 2.089 |
Fosl2
(Fra-2)
|

|
|
| GGAAUGU | 2.073 | mmu-miR-1a mmu-miR-206 |

|

|
| UGAAAUG | 2.073 | mmu-miR-203 |

|

|
| BACH2.p2 | 2.073 |
Bach2
|

|
|
| AAAGUGC | 2.072 | mmu-miR-17 mmu-miR-20a mmu-miR-20b mmu-miR-93 mmu-miR-106a mmu-miR-106b |

|

|
| GCAGCAU | 2.059 | mmu-miR-103 mmu-miR-107 |

|

|
| GUAAACA | 2.056 | mmu-miR-30a mmu-miR-30b mmu-miR-30c mmu-miR-30d mmu-miR-30e mmu-miR-384-5p |

|
|
| ACAUUCA | 2.053 | mmu-miR-181a mmu-miR-181b mmu-miR-181c mmu-miR-181d |

|

|
| SOX2.p2 | 2.047 |
Sox2
(ysb, Sox-2, lcc)
|

|
|
| CAGCAGG | 2.039 | mmu-miR-214 mmu-miR-761 |

|

|
| POU1F1.p2 | 2.034 |
Pou1f1
(Pit1-rs1, Hmp1, GHF-1, Pit1, Pit-1)
|
|
|
| CRX.p2 | 2.032 |
Crx
(Crx1)
|

|
|
| GGAAGAC | 2.016 | mmu-miR-7a mmu-miR-7b |

|

|
| HMGA1,2.p2 | 1.985 |
Hmga1
(Hmga1b, Hmgi, HMGI(Y), Hmga1a, HMG-I(Y), HMGY, Hmgiy)
Hmga2 (HMGI-C, Hmgic) |

|
|
| DBP.p2 | 1.970 |
Dbp
|

|
|
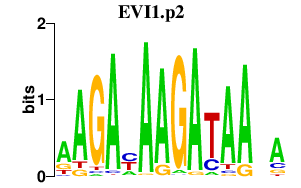
| EVI1.p2 | 1.966 |
Mecom
(MDS1-EVI1, Prdm3, Jbo, Mds1, ZNFPR1B1, Evi-1, Evi1)
|

|
|
| SOX5.p2 | 1.962 |
Sox5
|

|

|
| AGCAGCA | 1.950 | mmu-miR-15a mmu-miR-15b mmu-miR-16 mmu-miR-195 mmu-miR-322 mmu-miR-497 mmu-miR-1907 |

|

|
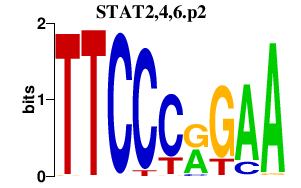
| STAT2,4,6.p2 | 1.942 |
Stat2
Stat4 Stat6 |
|
|
| CCAGUGU | 1.940 | mmu-miR-199a-5p |

|

|
| GGCUCAG | 1.930 | mmu-miR-24 |

|

|
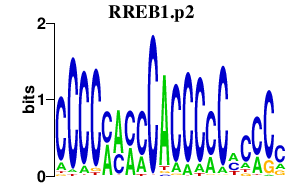
| RREB1.p2 | 1.920 |
Rreb1
|

|
|
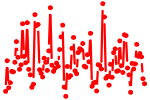
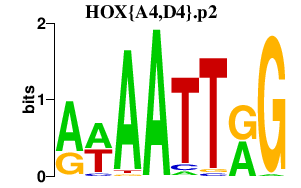
| HOX{A4,D4}.p2 | 1.916 |
Hoxa4
(Hox-1.4)
Hoxd4 (Hox-4.2, Hox-5.1) |
|
|
| AUGGCUU | 1.911 | mmu-miR-135a mmu-miR-135b |
|

|
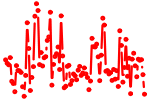
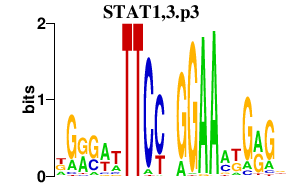
| STAT1,3.p3 | 1.909 |
Stat1
Stat3 (Aprf) |

|
|
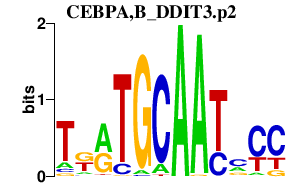
| CEBPA,B_DDIT3.p2 | 1.905 |
Ddit3
(CHOP-10, C/EBP homoologous protein 10, chop, gadd153, CHOP10)
Cebpa (C/ebpalpha, Cebp, C/EBP alpha) Cebpb (CRP2, C/EBPbeta, Nfil6, LAP, C/EBP BETA, LIP, NF-M, IL-6DBP, NF-IL6) |
|
|
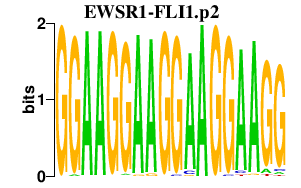
| EWSR1-FLI1.p2 | 1.903 |
Fli1
(EWSR2, Sic1, SIC-1, Fli-1)
Ewsr1 (Ews) |

|
|
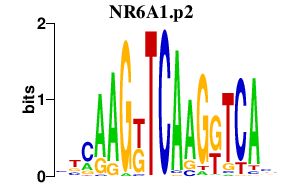
| NR6A1.p2 | 1.902 |
Nr6a1
(NCNF, Gcnf)
|

|
|
| TFAP2B.p2 | 1.897 |
Tcfap2b
(AP-2(beta))
|

|
|
| AIRE.p2 | 1.880 |
Aire
|

|
|
| GCUGGUG | 1.869 | mmu-miR-138 |

|

|
| GUGCAAA | 1.867 | mmu-miR-19a mmu-miR-19b |

|

|
| PAX2.p2 | 1.856 |
Pax2
(Pax-2, Opdc)
|

|
|
| GTF2A1,2.p2 | 1.856 |
Gtf2a1
(37kDa, 19kDa, TfIIAa/b)
Gtf2a2 (TfIIg, 12kDa) |

|
|
| CDX1,2,4.p2 | 1.844 |
Cdx1
(Cdx, Cdx-1)
Cdx2 (Cdx-2) Cdx4 (Cdx-3, Cdx-4, Cdx3) |

|
|
| ESR1.p2 | 1.836 |
Esr1
(ER-alpha, ERalpha, ESR, ER[a], Nr3a1, ERa, Estra, Estr)
|

|
|
| NFE2.p2 | 1.831 |
Nfe2
(p45nf-e2, p45NFE2, p45, NF-E2)
|

|
|
| GFI1B.p2 | 1.823 |
Gfi1b
(Gfi-1B)
|

|
|
| AAGUGCU | 1.817 | mmu-miR-291a-3p mmu-miR-294 mmu-miR-295 mmu-miR-302a mmu-miR-302b mmu-miR-302d |

|
|
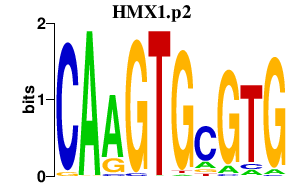
| HMX1.p2 | 1.813 |
Hmx1
(Nkx5-3)
|

|
|
| AUGGCAC | 1.809 | mmu-miR-183 |

|

|
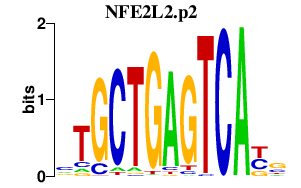
| NFE2L2.p2 | 1.806 |
Nfe2l2
(Nrf2)
|

|
|
| GAGGUAG | 1.806 | mmu-let-7a mmu-let-7b mmu-let-7c mmu-let-7d mmu-let-7e mmu-let-7f mmu-let-7g mmu-let-7i mmu-miR-98 mmu-miR-1961 |
|

|
| UUUUUGC | 1.805 | mmu-miR-129-5p |

|

|
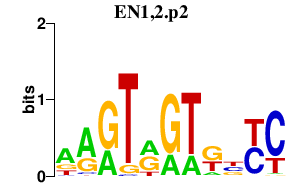
| EN1,2.p2 | 1.801 |
En1
(engrailed-1, Mo-en.1, En-1)
En2 (En-2) |

|
|
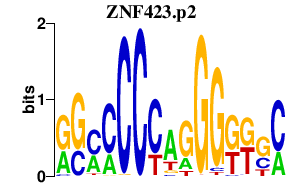
| ZNF423.p2 | 1.797 |
Zfp423
(ataxia1, Ebfaz, Zfp104, Roaz)
|

|
|
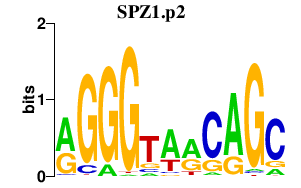
| SPZ1.p2 | 1.795 |
Spz1
|

|
|
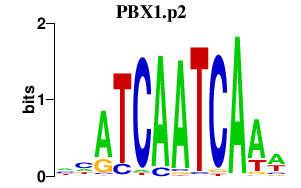
| PBX1.p2 | 1.782 |
Pbx1
(Pbx-1)
|

|
|
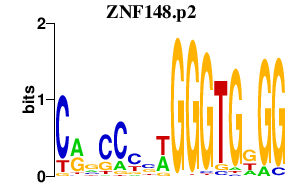
| ZNF148.p2 | 1.768 |
Zfp148
(beta enolase repressor factor 1, ZBP-89, BFCOL1, BERF-1)
|

|
|
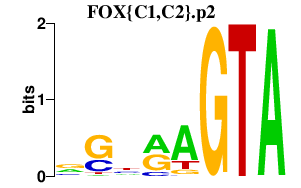
| FOX{C1,C2}.p2 | 1.763 |
Foxc1
(Mf1, frkhda, fkh-1, Mf4, FREAC3, fkh1, Fkh1)
Foxc2 (Hfhbf3, Fkh14, MFH-1, Mfh1) |
|
|
| NKX6-1,2.p2 | 1.763 |
Nkx6-1
(NKX6A, Nkx6.1)
Nkx6-2 (Nkx6.2, Gtx) |
|
|
| EBF1.p2 | 1.742 |
Ebf1
(Olf-1, Olf1, O/E-1)
|

|
|
| UGCAUAG | 1.739 | mmu-miR-153 |

|

|
| CREB1.p2 | 1.734 |
Creb1
(Creb-1, Creb)
|

|
|
| PAX4.p2 | 1.724 |
Pax4
(Pax-4)
|

|
|
| ZBTB16.p2 | 1.701 |
Zbtb16
(Zfp145, PLZF, Green's luxoid)
|

|
|
| NKX3-2.p2 | 1.693 |
Nkx3-2
(Nkx-3.2, Bapx1)
|

|
|
| POU5F1_SOX2{dimer}.p2 | 1.671 |
Pou5f1
(Otf3-rs7, Oct3/4, Otf-4, Oct-3, Oct4, Otf3, Oct-3/4, Otf3g, Otf-3, Otf4, Oct-4)
Sox2 (ysb, Sox-2, lcc) |
|
|
| TFAP4.p2 | 1.658 |
Tcfap4
(bHLHc41, Tfap4, AP-4)
|

|
|
| FOXQ1.p2 | 1.639 |
Foxq1
(Hfh1l, HFH-1, Hfh1, sa)
|

|
|
| SMAD1..7,9.p2 | 1.621 |
Smad5
(MusMLP, Madh5, Smad 5)
Smad1 (Smad 1, Madh1, Madr1) Smad3 (Smad 3, Madh3) Smad4 (Smad 4, DPC4, Dpc4, Madh4, D18Wsu70e) Smad2 (Smad 2, Madh2, Madr2) Smad6 (Smad 6, Madh6) Smad7 (Madh7) Smad9 (SMAD8A, SMAD8B, Madh9, MADH6) |

|
|
| NR3C1.p2 | 1.604 |
Nr3c1
(glucocorticoid receptor, GR, Grl1, Grl-1)
|

|
|
| POU3F1..4.p2 | 1.602 |
Pou3f1
(Tst-1, Test1, Scip, Oct6, Oct-6, Tst1, Otf6)
Pou3f3 (Brn1, Brn-1, Otf8) Pou3f2 (Otf7, Brn2, Brn-2) Pou3f4 (BRN-4, Otf9, Brn4) |

|
|
| MYBL2.p2 | 1.600 |
Mybl2
(Bmyb, B-Myb)
|

|
|
| IKZF2.p2 | 1.579 |
Ikzf2
(Zfpn1a2, Helios)
|

|
|
| AGCUUAU | 1.578 | mmu-miR-21 mmu-miR-590-5p |

|

|
| GUCAGUU | 1.571 | mmu-miR-223 |

|

|
| NFIL3.p2 | 1.566 |
Nfil3
(E4BP4)
|

|
|
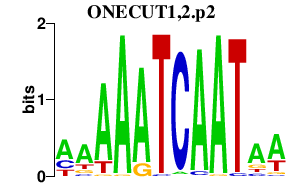
| ONECUT1,2.p2 | 1.561 |
Onecut2
(OC-2, Oc2)
Onecut1 (OC-1, HNF6, Hfh12, Hnf6, Oc1, D9Ertd423e) |

|
|
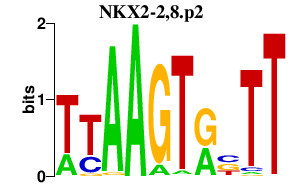
| NKX2-2,8.p2 | 1.556 |
Nkx2-2
(Nkx2.2, tinman, Nkx-2.2)
Nkx2-9 (tinman, Nkx-2.9, Nkx2.9) |

|
|
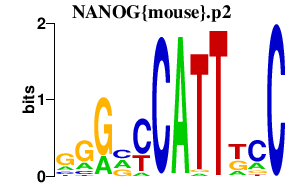
| NANOG{mouse}.p2 | 1.552 |
Nanog
(ecat4, ENK)
|

|
|
| PAX5.p2 | 1.546 |
Pax5
(Pax-5, EBB-1)
|
|
|
| EOMES.p2 | 1.544 |
Eomes
(Tbr2)
|

|
|
| HOX{A5,B5}.p2 | 1.487 |
Hoxb5
(Hox-2.1)
Hoxa5 (Hox-1.3) |

|
|
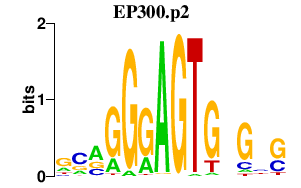
| EP300.p2 | 1.487 |
Ep300
(KAT3B, p300)
|
|
|
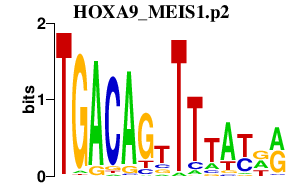
| HOXA9_MEIS1.p2 | 1.474 |
Meis1
Hoxa9 (Hox-1.7, D6a9) |
|
|
| LHX3,4.p2 | 1.469 |
Lhx3
(P-LIM, mLim-3, Lim3)
Lhx4 (Gsh4, Gsh-4) |
|
|
| RXRA_VDR{dimer}.p2 | 1.458 |
Vdr
(Nr1i1)
|

|

|
| UCACAUU | 1.452 | mmu-miR-23a mmu-miR-23b |

|

|
| FOXN1.p2 | 1.444 |
Foxn1
(Hfh11, D11Bhm185e, whn)
|

|
|
| FOXO1,3,4.p2 | 1.436 |
Foxo1
(FKHR, Foxo1a, Fkhr1, Afxh)
Foxo4 (Mllt7, Foxo4, afx, Afxh) Foxo3 (FKHRL1, Foxo3a, Fkhr2) |

|
|
| ALX1.p2 | 1.416 |
Alx1
(Cart1)
|
|
|
| NKX3-1.p2 | 1.408 |
Nkx3-1
(Nkx-3.1, bagpipe, NKX3.1, NKX3A, Bax)
|
|
|
| NFIX.p2 | 1.408 |
Nfix
|

|
|
| FOXP3.p2 | 1.399 |
Foxp3
(scurfin, JM2)
|

|
|
| GCUACAU | 1.397 | mmu-miR-221 mmu-miR-222 mmu-miR-1928 |

|

|
| NKX2-1,4.p2 | 1.396 |
Nkx2-1
(T/EBP, thyroid transcription factor-1, Titf1, thyroid-specific enhancer-binding protein, tinman, Ttf-1)
Nkx2-4 (Nkx-2.4, tinman) |

|
|
| UUGGCAC | 1.389 | mmu-miR-96 |

|

|
| CDC5L.p2 | 1.372 |
Cdc5l
(PCDC5RP)
|

|
|
| AGCUGCC | 1.365 | mmu-miR-22 |

|

|
| AGUGGUU | 1.344 | mmu-miR-140 mmu-miR-876-3p |

|

|
| NANOG.p2 | 1.329 |
Nanog
(ecat4, ENK)
|

|

|
| SRY.p2 | 1.276 |
Sry
(Tdf, Tdy)
|

|
|
| CUCCCAA | 1.276 | mmu-miR-150 mmu-miR-5127 |

|

|
| AGUGCAA | 1.263 | mmu-miR-130a mmu-miR-130b mmu-miR-301a mmu-miR-301b mmu-miR-721 |

|

|
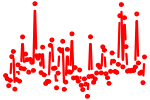
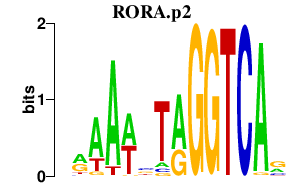
| RORA.p2 | 1.260 |
Rora
(Nr1f1, tmgc26)
|
|
|
| AGCAGCG | 1.243 | mmu-miR-503 |

|

|
| UCAAGUA | 1.225 | mmu-miR-26a mmu-miR-26b |

|

|
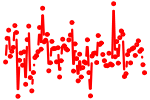
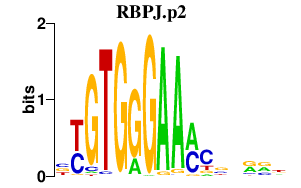
| RBPJ.p2 | 1.216 |
Rbpj
(RBPjk, RBP-J kappa, Rbpsuh, Igkjrb, CBF1, Igkrsbp)
|
|
|
| CAGUGCA | 1.209 | mmu-miR-148a mmu-miR-148b mmu-miR-152 |

|
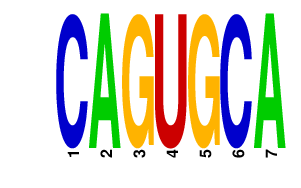
|
| UCCCUUU | 1.208 | mmu-miR-204 mmu-miR-211 |

|
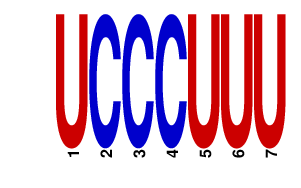
|
| NKX2-3_NKX2-5.p2 | 1.206 |
Nkx2-5
(Nkx-2.5, Nkx2.5, Csx, tinman)
Nkx2-3 (Nkx-2.3, tinman, Nkx2.3) |
|
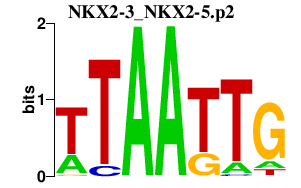
|
| AACAGUC | 1.191 | mmu-miR-132 mmu-miR-212-3p |
|

|
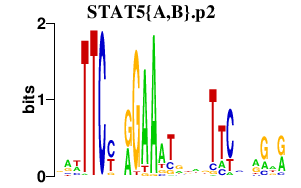
| STAT5{A,B}.p2 | 1.188 |
Stat5b
Stat5a (STAT5) |

|
|
| CCUUCAU | 1.186 | mmu-miR-205 |

|

|
| GUAACAG | 1.179 | mmu-miR-194 |
|

|
| AUUGCAC | 1.176 | mmu-miR-25 mmu-miR-32 mmu-miR-92a mmu-miR-92b mmu-miR-363-3p mmu-miR-367 |

|

|
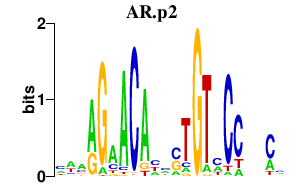
| AR.p2 | 1.138 |
Ar
|

|
|
| UUGUUCG | 1.130 | mmu-miR-375 |

|

|
| CCAGCAU | 1.113 | mmu-miR-338-3p |

|

|
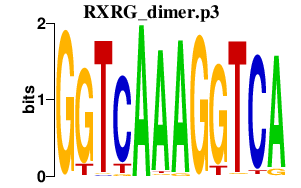
| RXRG_dimer.p3 | 1.108 |
Nr1h2
(LXRbeta, RIP15, Unr2, LXRB)
Rxra (RXR alpha 1, RXRalpha1) Pparg (Ppar-gamma2, PPAR-gamma, PPARgamma, PPARgamma2, Nr1c3) Ppara (PPAR-alpha, PPARalpha, Nr1c1, Ppar) Rxrb (Nr2b2, H-2RIIBP, RCoR-1, Rub) Rxrg (Nr2b3) |

|
|
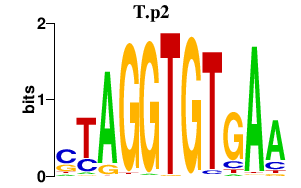
| T.p2 | 1.101 |
T
(T1, Bra)
|

|
|
| UUGGCAA | 1.093 | mmu-miR-182 |

|

|
| GAUCAGA | 1.091 | mmu-miR-383 |

|

|
| AAGGUGC | 1.088 | mmu-miR-18a mmu-miR-18b |
|

|
| UGCAUUG | 1.084 | mmu-miR-33 |
|

|
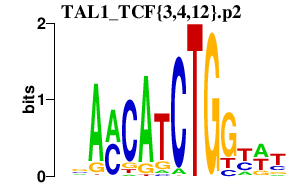
| TAL1_TCF{3,4,12}.p2 | 1.079 |
Tcf3
(ALF2, E12, bHLHb21, Tcfe2a, A1, E47, Pan1, E2A, Pan2)
Tcf4 (E2.2, bHLHb19, SEF-2, TFE, ME2, ITF-2b, ASP-I2, MITF-2A, SEF2-1, E2-2, ITF-2, MITF-2B) Tal1 (bHLHa17, Scl, SCL/tal-1) Tcf12 (HTF4, bHLHb20, HEB, HTF-4, REB, ALF1, HEBAlt, ME1) |
|
|
| ACAGUAC | 1.072 | mmu-miR-101a mmu-miR-101b |

|

|
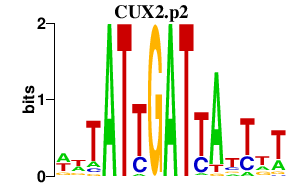
| CUX2.p2 | 1.065 |
Cux2
(ENSMUSG00000072641, Cux-2, Cutl2, Cux2)
|

|
|
| GGCAAGA | 1.018 | mmu-miR-31 |

|

|
| AAUGCCC | 1.004 | mmu-miR-365 |

|

|
| GAUUGUC | 1.001 | mmu-miR-219-5p |
|

|
| UAAGACG | 0.995 | mmu-miR-208a-3p mmu-miR-208b |

|

|
| PAX6.p2 | 0.989 |
Pax6
(Pax-6, Dickie's small eye, AEY11, Dey, Gsfaey11)
|

|

|
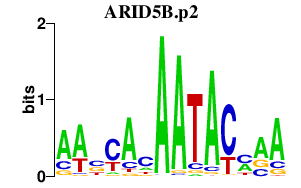
| ARID5B.p2 | 0.983 |
Arid5b
(Mrf2, Mrf2alpha, Desrt, Mrf2beta)
|

|
|
| UAAGACU | 0.955 | mmu-miR-499 |

|

|
| TP53.p2 | 0.936 |
Trp53
(p44, p53)
|
|
|
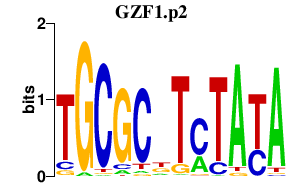
| GZF1.p2 | 0.931 |
Gzf1
(Zfp336)
|

|
|
| CACAGUG | 0.918 | mmu-miR-128 |

|

|
| UAAUGCU | 0.870 | mmu-miR-155 |

|

|
| AGGUAGU | 0.853 | mmu-miR-196a mmu-miR-196b |

|

|
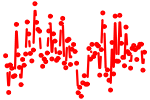
| ACUGGCC | 0.840 | mmu-miR-193 mmu-miR-193b |
|

|
| ACAGUAU | 0.837 | mmu-miR-144 |

|

|
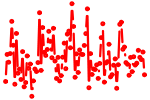
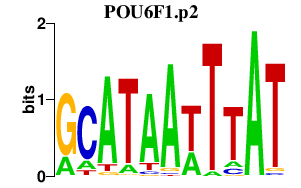
| POU6F1.p2 | 0.830 |
Pou6f1
(Emb, cns-1)
|
|
|
| ACCCUGU | 0.811 | mmu-miR-10a mmu-miR-10b |

|

|
| UCACAGU | 0.790 | mmu-miR-27a mmu-miR-27b |

|

|
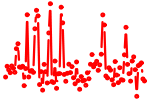
| ACUGCAU | 0.786 | mmu-miR-217 |
|

|
| AUGACAC | 0.779 | mmu-miR-425 mmu-miR-489 |
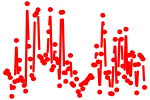
|

|
| GAGAACU | 0.754 | mmu-miR-146a mmu-miR-146b |

|
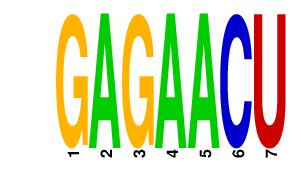
|
| CUACAGU | 0.729 | mmu-miR-139-5p |

|

|
| PPARG.p2 | 0.710 |
Pparg
(Ppar-gamma2, PPAR-gamma, PPARgamma, PPARgamma2, Nr1c3)
|

|
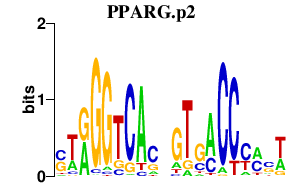
|
| MSX1,2.p2 | 0.675 |
Msx1
(msh, Hox7.1, Hox7, muscle-segment homeobox, Hox-7)
Msx2 (Hox-8, Hox8, Hox8.1) |

|

|
| VSX1,2.p2 | 0.664 |
Vsx2
(Hox10, Chx10, Hox-10)
Vsx1 (CHX10-like) |

|
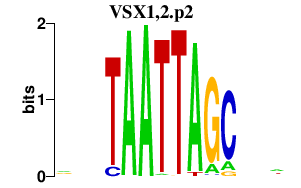
|
| GAGAUGA | 0.660 | mmu-miR-143 |

|

|
| AAUCUCU | 0.596 | mmu-miR-216b |

|

|
| PDX1.p2 | 0.545 |
Pdx1
(Ipf1, IPF-1, pdx-1, STF-1, Mody4, IDX-1)
|

|
|
| GGACGGA | 0.477 | mmu-miR-184 |

|
|
| AACGGAA | 0.471 | mmu-miR-191 |

|
|
| CGUGUCU | 0.449 | mmu-miR-187 |

|

|
| GGAGUGU | 0.449 | mmu-miR-122 |

|
|
| AAUCUCA | 0.426 | mmu-miR-216a |

|

|
| AACCUGG | 0.365 | mmu-miR-490-3p |
|

|
| GAUAUGU | 0.360 | mmu-miR-190 mmu-miR-190b |

|

|
| CGUACCG | 0.336 | mmu-miR-126-3p |

|

|
| UGACCUA | 0.313 | mmu-miR-192 mmu-miR-215 |
|

|
| UGUGCGU | 0.288 | mmu-miR-210 |
|

|
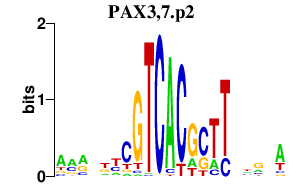
| PAX3,7.p2 | 0.177 |
Pax7
(Pax-7)
Pax3 (Pax-3) |

|
|
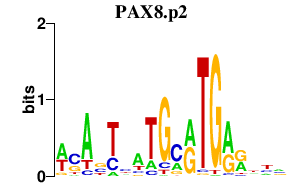
| PAX8.p2 | 0.174 |
Pax8
(Pax-8)
|
|
|
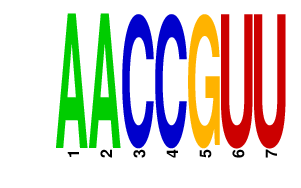
| AACCGUU | 0.102 | mmu-miR-451 |
|
|
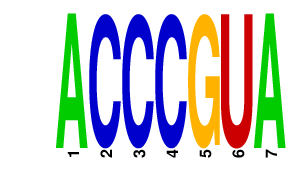
| ACCCGUA | 0.024 | mmu-miR-99a mmu-miR-99b mmu-miR-100 |
|
|
All samples sorted alphabetically
| Your email address: | |
| Project name: |