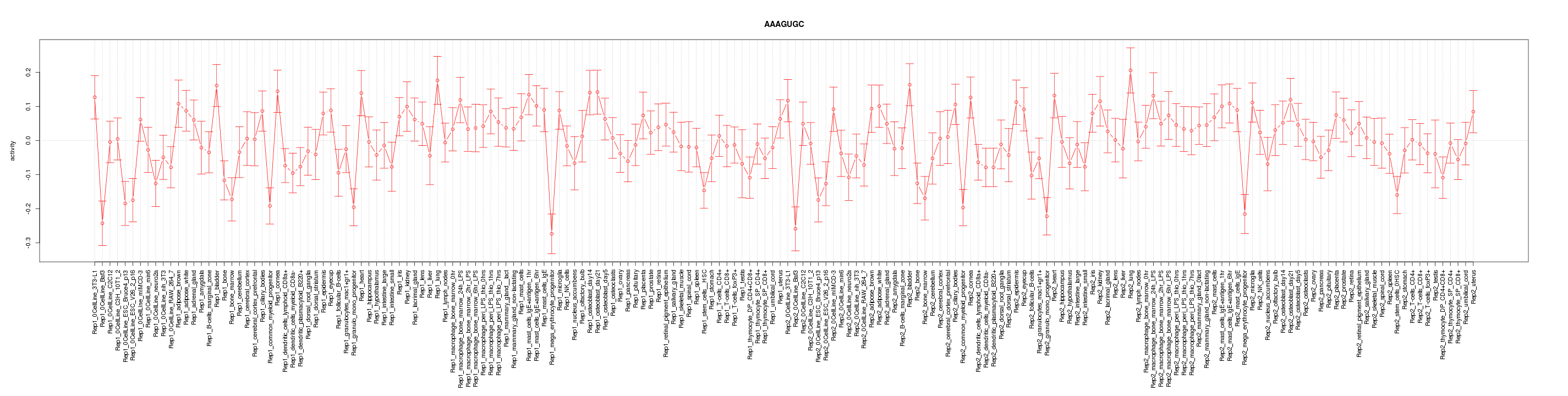
|
chr3_-_57651621
|
17.231
|
NM_019410
|
Pfn2
|
profilin 2
|
|
chr5_+_104888468
|
15.099
|
NM_008861
|
Pkd2
|
polycystic kidney disease 2
|
|
chrX_+_136145155
|
14.313
|
NM_023270
|
Rnf128
|
ring finger protein 128
|
|
chr15_+_6249788
|
13.460
|
NM_001008702
|
Dab2
|
disabled homolog 2 (Drosophila)
|
|
chr15_-_99705812
|
12.675
|
NM_001113545
|
Lima1
|
LIM domain and actin binding 1
|
|
chr3_+_121426422
|
12.501
|
NM_010171
|
F3
|
coagulation factor III
|
|
chr10_-_8605668
|
12.434
|
NM_175155
|
Sash1
|
SAM and SH3 domain containing 1
|
|
chr6_+_34659440
|
12.316
|
NM_145575
|
Cald1
|
caldesmon 1
|
|
chr15_-_99650502
|
12.095
|
NM_023063
|
Lima1
|
LIM domain and actin binding 1
|
|
chr11_-_118216689
|
11.885
|
NM_011594
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr1_-_74170987
|
11.701
|
NM_027884
|
Tns1
|
tensin 1
|
|
chr8_-_26127376
|
11.428
|
NM_007404
|
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma)
|
|
chr7_-_125635435
|
10.947
|
NM_001033380
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chr3_-_57379831
|
10.883
|
NM_001168281
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr5_+_17080553
|
10.744
|
NM_013657
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr7_-_150927867
|
10.529
|
NM_024289
|
Osbpl5
|
oxysterol binding protein-like 5
|
|
chr5_+_75552144
|
10.447
|
NM_001083316
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr9_-_123760686
|
10.372
|
NM_148925
|
Fyco1
|
FYVE and coiled-coil domain containing 1
|
|
chr15_+_6336747
|
10.316
|
NM_001037905
NM_001102400
NM_023118
|
Dab2
|
disabled homolog 2 (Drosophila)
|
|
chr17_+_29227908
|
10.101
|
NM_001111099
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21)
|
|
chr8_+_63133900
|
10.091
|
NM_001177881
|
Mfap3l
|
microfibrillar-associated protein 3-like
|
|
chr3_-_57379600
|
9.865
|
NM_133784
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr17_+_29230715
|
9.752
|
NM_007669
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21)
|
|
chr3_-_55987612
|
9.626
|
NM_030595
|
Nbea
|
neurobeachin
|
|
chr14_-_55196138
|
9.609
|
NM_010590
|
Jub
|
ajuba
|
|
chr17_+_57388873
|
9.404
|
NM_134125
|
Trip10
|
thyroid hormone receptor interactor 10
|
|
chr16_-_85173944
|
9.140
|
NM_001198823
NM_001198824
NM_001198825
NM_001198826
NM_007471
|
App
|
amyloid beta (A4) precursor protein
|
|
chr6_+_116288075
|
9.069
|
NM_027920
|
March8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr11_-_78349594
|
9.015
|
NM_001159392
NM_009395
|
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr8_+_63134353
|
8.858
|
NM_001177882
|
Mfap3l
|
microfibrillar-associated protein 3-like
|
|
chr16_-_21694608
|
8.727
|
NM_001001881
|
2510009E07Rik
|
RIKEN cDNA 2510009E07 gene
|
|
chr17_+_14416508
|
8.706
|
NM_022315
|
Smoc2
|
SPARC related modular calcium binding 2
|
|
chr9_-_116084395
|
8.591
|
NM_009371
NM_029575
|
Tgfbr2
|
transforming growth factor, beta receptor II
|
|
chr5_+_74591336
|
8.359
|
NM_026878
|
Rasl11b
|
RAS-like, family 11, member B
|
|
chr17_+_44215761
|
8.338
|
NM_032003
NM_001168620
|
Enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5
|
|
chr7_-_152125823
|
8.259
|
NM_007631
|
Ccnd1
|
cyclin D1
|
|
chr2_+_59450080
|
8.227
|
NM_198294
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr9_-_51976215
|
8.204
|
NM_001162921
|
Zc3h12c
|
zinc finger CCCH type containing 12C
|
|
chr5_-_76733540
|
8.165
|
NM_007715
|
Clock
|
circadian locomotor output cycles kaput
|
|
chr1_-_82583400
|
8.058
|
NM_007735
|
Col4a4
|
collagen, type IV, alpha 4
|
|
chr8_+_63111621
|
7.956
|
NM_027756
|
Mfap3l
|
microfibrillar-associated protein 3-like
|
|
chr6_-_85324557
|
7.877
|
NM_001003955
NM_177466
|
Rab11fip5
|
RAB11 family interacting protein 5 (class I)
|
|
chr12_+_9581218
|
7.737
|
NM_011859
|
Osr1
|
odd-skipped related 1 (Drosophila)
|
|
chr4_-_138648762
|
7.688
|
NM_008675
|
Nbl1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr5_-_37380147
|
7.476
|
NM_011716
|
Wfs1
|
Wolfram syndrome 1 homolog (human)
|
|
chr1_-_90598670
|
7.452
|
NM_177305
|
Arl4c
|
ADP-ribosylation factor-like 4C
|
|
chr7_-_112781902
|
7.422
|
NM_018880
|
Trim3
|
tripartite motif-containing 3
|
|
chr16_+_58408609
|
7.400
|
NM_028523
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr9_-_63869857
|
7.097
|
NM_008542
|
Smad6
|
MAD homolog 6 (Drosophila)
|
|
chr11_+_105451296
|
7.056
|
NM_181071
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr13_-_13485585
|
6.955
|
NM_031999
|
Gpr137b
|
G protein-coupled receptor 137B
|
|
chr11_-_80593259
|
6.914
|
NM_177390
|
Myo1d
|
myosin ID
|
|
chr4_-_82151180
|
6.760
|
NM_001113209
NM_001113210
NM_008687
|
Nfib
|
nuclear factor I/B
|
|
chr6_-_89312545
|
6.718
|
NM_008881
|
Plxna1
|
plexin A1
|
|
chr17_+_15080188
|
6.712
|
NM_001083880
NM_001083881
NM_001083882
NM_026451
|
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene
|
|
chr17_-_15963372
|
6.706
|
NM_178615
|
Rgmb
|
RGM domain family, member B
|
|
chr5_-_148537561
|
6.642
|
NM_010228
|
Flt1
|
FMS-like tyrosine kinase 1
|
|
chr4_-_57314708
|
6.481
|
NM_011207
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr3_+_7366586
|
6.466
|
NM_008862
|
Pkia
|
protein kinase inhibitor, alpha
|
|
chr10_-_7500674
|
6.450
|
NM_145418
|
BC013529
|
cDNA sequence BC013529
|
|
chr12_+_71926429
|
6.443
|
NM_028127
|
Frmd6
|
FERM domain containing 6
|
|
chr2_+_91490266
|
6.413
|
NM_001145902
|
Arhgap1
|
Rho GTPase activating protein 1
|
|
chr14_-_37781943
|
6.398
|
NM_027045
|
Gcap14
|
granule cell antiserum positive 14
|
|
chr3_+_104591946
|
6.369
|
NM_007484
|
Rhoc
|
ras homolog gene family, member C
|
|
chr17_+_78599556
|
6.347
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr10_-_66559568
|
6.344
|
NM_178606
|
Reep3
|
receptor accessory protein 3
|
|
chr19_+_27291507
|
6.328
|
NM_001161420
NM_013703
|
Vldlr
|
very low density lipoprotein receptor
|
|
chr9_+_122260733
|
6.324
|
NM_026179
|
Abhd5
|
abhydrolase domain containing 5
|
|
chr14_+_117324518
|
6.275
|
NM_001079844
NM_011821
|
Gpc6
|
glypican 6
|
|
chr8_+_36438761
|
6.258
|
NM_177741
|
Ppp1r3b
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr19_+_38911056
|
6.180
|
NM_145952
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12
|
|
chr5_+_89316245
|
6.122
|
NM_001136260
NM_001197147
NM_018760
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4
|
|
chr2_-_166888514
|
6.088
|
NM_001033196
|
Znfx1
|
zinc finger, NFX1-type containing 1
|
|
chr16_-_85901362
|
6.079
|
NM_011782
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2)
|
|
chr17_-_14831262
|
6.030
|
NM_011581
|
Thbs2
|
thrombospondin 2
|
|
chr4_-_53172748
|
5.949
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr3_+_38785861
|
5.946
|
NM_183221
|
Fat4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr5_+_75548309
|
5.937
|
NM_011058
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr19_-_34821529
|
5.703
|
NM_172838
|
Slc16a12
|
solute carrier family 16 (monocarboxylic acid transporters), member 12
|
|
chr13_+_113254277
|
5.675
|
NM_010560
|
Il6st
|
interleukin 6 signal transducer
|
|
chr8_+_85387234
|
5.644
|
NM_177378
|
Rnf150
|
ring finger protein 150
|
|
chr17_+_72131286
|
5.615
|
NM_030179
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr15_-_67008433
|
5.424
|
NM_009177
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr6_+_83745142
|
5.385
|
NM_001164187
|
Nagk
|
N-acetylglucosamine kinase
|
|
chr6_+_114593104
|
5.378
|
NM_028835
|
Atg7
|
autophagy-related 7 (yeast)
|
|
chr16_-_59555577
|
5.335
|
NM_174848
|
Crybg3
|
beta-gamma crystallin domain containing 3
|
|
chr6_-_72566993
|
5.333
|
NM_009443
|
Tgoln1
|
trans-golgi network protein
|
|
chr1_+_82583495
|
5.314
|
NM_007734
|
Col4a3
|
collagen, type IV, alpha 3
|
|
chr10_+_69707965
|
5.282
|
NM_025807
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9
|
|
chr19_+_7568529
|
5.236
|
NM_001163505
|
Atl3
|
atlastin GTPase 3
|
|
chr11_-_70832737
|
5.225
|
NM_033562
|
Derl2
|
Der1-like domain family, member 2
|
|
chr19_+_29441927
|
5.215
|
NM_021893
|
Cd274
|
CD274 antigen
|
|
chr5_+_123157541
|
5.169
|
NM_011026
|
P2rx4
|
purinergic receptor P2X, ligand-gated ion channel 4
|
|
chr1_+_12682510
|
5.168
|
NM_001198565
NM_001198566
|
Sulf1
|
sulfatase 1
|
|
chr4_-_154596628
|
5.154
|
NM_011385
|
Ski
|
ski sarcoma viral oncogene homolog (avian)
|
|
chr6_-_84543739
|
5.145
|
NM_175475
NM_001177713
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1
|
|
chr8_+_109459623
|
5.107
|
NM_009229
|
Sntb2
|
syntrophin, basic 2
|
|
chr11_-_40568710
|
5.100
|
NM_009831
|
Ccng1
|
cyclin G1
|
|
chr17_+_24686778
|
4.987
|
NM_013630
|
Pkd1
|
polycystic kidney disease 1 homolog
|
|
chr14_+_31528800
|
4.982
|
NM_001166532
|
Sfmbt1
|
Scm-like with four mbt domains 1
|
|
chr14_-_67487226
|
4.894
|
NM_009955
|
Dpysl2
|
dihydropyrimidinase-like 2
|
|
chr15_-_60656465
|
4.889
|
NM_001162926
|
Fam84b
|
family with sequence similarity 84, member B
|
|
chr7_-_17200285
|
4.880
|
NM_172739
|
Grlf1
|
glucocorticoid receptor DNA binding factor 1
|
|
chr11_-_98448496
|
4.876
|
NM_025661
|
Ormdl3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr1_+_193541845
|
4.855
|
NM_172266
|
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr12_+_81891479
|
4.812
|
NM_178682
|
4933426M11Rik
|
RIKEN cDNA 4933426M11 gene
|
|
chr17_-_46162111
|
4.787
|
NM_001110266
NM_001110267
NM_001110268
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr14_-_37748753
|
4.777
|
NM_028407
|
Gcap14
|
granule cell antiserum positive 14
|
|
chr5_+_114185777
|
4.743
|
NM_001010825
|
Ficd
|
FIC domain containing
|
|
chr1_-_174228105
|
4.714
|
NM_178405
|
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr15_+_95621222
|
4.697
|
NM_175344
|
Ano6
|
anoctamin 6
|
|
chr1_+_193542261
|
4.670
|
NM_001134829
|
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr1_-_51535232
|
4.652
|
NM_028696
|
Obfc2a
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr6_-_72566691
|
4.648
|
NM_009444
|
Tgoln1
Tgoln2
|
trans-golgi network protein
trans-golgi network protein 2
|
|
chr4_+_136025675
|
4.641
|
NM_024452
|
Luzp1
|
leucine zipper protein 1
|
|
chr4_+_138011061
|
4.500
|
NM_025451
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr3_+_96361879
|
4.468
|
NM_001009935
NM_023719
|
Txnip
|
thioredoxin interacting protein
|
|
chr12_+_8928112
|
4.427
|
NM_008640
|
Laptm4a
|
lysosomal-associated protein transmembrane 4A
|
|
chr7_-_74904546
|
4.396
|
NM_183312
NM_201639
NM_207663
|
Synm
|
synemin, intermediate filament protein
|
|
chr9_-_123760992
|
4.373
|
NM_001110253
|
Fyco1
|
FYVE and coiled-coil domain containing 1
|
|
chr17_-_46169322
|
4.357
|
NM_001025250
NM_001025257
NM_009505
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr19_+_23215704
|
4.324
|
NM_010638
|
Klf9
|
Kruppel-like factor 9
|
|
chr9_-_101154160
|
4.312
|
NM_001161362
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr12_-_55304834
|
4.294
|
NM_028133
|
Egln3
|
EGL nine homolog 3 (C. elegans)
|
|
chr11_+_69579376
|
4.272
|
NM_029348
|
Zbtb4
|
zinc finger and BTB domain containing 4
|
|
chr4_+_86520317
|
4.227
|
NM_139306
|
Acer2
|
alkaline ceramidase 2
|
|
chr8_-_88364846
|
4.202
|
NM_028007
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr10_+_95403283
|
4.199
|
NM_001001932
|
Eea1
|
early endosome antigen 1
|
|
chr12_-_45311020
|
4.179
|
NM_013760
|
Dnajb9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr19_-_47766512
|
4.176
|
NM_007732
|
Col17a1
|
collagen, type XVII, alpha 1
|
|
chr11_-_68200284
|
4.133
|
NM_008744
|
Ntn1
|
netrin 1
|
|
chr18_+_12492494
|
4.120
|
NM_010680
|
Lama3
|
laminin, alpha 3
|
|
chr1_+_12708582
|
4.102
|
NM_172294
|
Sulf1
|
sulfatase 1
|
|
chr6_-_137118238
|
4.096
|
NM_001164212
NM_001164214
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor
|
|
chr6_+_91937103
|
4.017
|
NM_172731
|
Fgd5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr11_-_69761489
|
4.007
|
NM_009204
|
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr6_-_94650118
|
3.997
|
NM_008377
|
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr5_-_140865121
|
3.939
|
NM_172277
|
Snx8
|
sorting nexin 8
|
|
chr1_-_77511653
|
3.932
|
NM_007936
|
Epha4
|
Eph receptor A4
|
|
chr15_+_85565935
|
3.917
|
NM_001113418
|
Ppara
|
peroxisome proliferator activated receptor alpha
|
|
chr14_-_32249046
|
3.917
|
NM_011894
|
Sh3bp5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr5_-_147032932
|
3.906
|
NM_028774
|
Rnf6
|
ring finger protein (C3H2C3 type) 6
|
|
chr1_+_59821480
|
3.869
|
NM_007561
|
Bmpr2
|
bone morphogenic protein receptor, type II (serine/threonine kinase)
|
|
chr15_+_85566122
|
3.866
|
NM_011144
|
Ppara
|
peroxisome proliferator activated receptor alpha
|
|
chr3_-_122322556
|
3.831
|
NM_001114665
NM_153118
|
Fnbp1l
|
formin binding protein 1-like
|
|
chr2_-_34227309
|
3.827
|
NM_016768
|
Pbx3
|
pre B-cell leukemia transcription factor 3
|
|
chr2_+_84890611
|
3.811
|
NM_001081260
|
Tnks1bp1
|
tankyrase 1 binding protein 1
|
|
chr4_-_147278927
|
3.807
|
NM_133201
|
Mfn2
|
mitofusin 2
|
|
chr3_-_38383688
|
3.770
|
NM_001167883
|
Ankrd50
|
ankyrin repeat domain 50
|
|
chr3_+_98186380
|
3.769
|
NM_172863
|
Zfp697
|
zinc finger protein 697
|
|
chr7_-_127039189
|
3.748
|
NM_001163703
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr8_+_11758322
|
3.738
|
NM_001113518
NM_017402
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7)
|
|
chr7_-_87547589
|
3.714
|
NM_011046
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr1_-_165243777
|
3.702
|
NM_001025570
NM_011127
NM_175686
|
Prrx1
|
paired related homeobox 1
|
|
chr6_-_128093543
|
3.675
|
NM_175414
|
Tspan9
|
tetraspanin 9
|
|
chr15_-_94420206
|
3.657
|
NM_008971
|
Twf1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr3_-_30868320
|
3.616
|
NM_001165954
NM_001165955
NM_001165956
NM_153421
|
Phc3
|
polyhomeotic-like 3 (Drosophila)
|
|
chr3_-_100914081
|
3.604
|
NM_011197
|
Ptgfrn
|
prostaglandin F2 receptor negative regulator
|
|
chr5_+_120120671
|
3.535
|
NM_011535
NM_198052
|
Tbx3
|
T-box 3
|
|
chr11_-_50105794
|
3.509
|
NM_175334
|
Maml1
|
mastermind like 1 (Drosophila)
|
|
chr7_-_87550315
|
3.505
|
NM_001081454
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr12_+_81745188
|
3.434
|
NM_026244
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr2_+_167906462
|
3.423
|
NM_021409
|
Pard6b
|
par-6 (partitioning defective 6) homolog beta (C. elegans)
|
|
chr15_-_99200868
|
3.416
|
NM_011711
|
Fmnl3
|
formin-like 3
|
|
chr7_-_51920870
|
3.405
|
NM_028021
|
Myh14
|
myosin, heavy polypeptide 14
|
|
chr5_-_64360058
|
3.398
|
NM_145923
|
Rell1
|
RELT-like 1
|
|
chr9_+_107465568
|
3.383
|
NM_019742
|
Tusc2
|
tumor suppressor candidate 2
|
|
chr5_-_36826896
|
3.378
|
NM_001170454
|
Tada2b
|
transcriptional adaptor 2B
|
|
chr3_-_10440061
|
3.375
|
NM_001127191
NM_029068
|
Snx16
|
sorting nexin 16
|
|
chr12_+_41172639
|
3.365
|
NM_172803
|
Dock4
|
dedicator of cytokinesis 4
|
|
chr13_-_34437042
|
3.349
|
NM_001033167
|
Slc22a23
|
solute carrier family 22, member 23
|
|
chr6_+_83745021
|
3.293
|
NM_019542
|
Nagk
|
N-acetylglucosamine kinase
|
|
chr14_+_28051224
|
3.282
|
NM_027871
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr12_-_99972803
|
3.281
|
NM_011877
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr12_-_99975614
|
3.252
|
NM_001146199
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr10_-_75807976
|
3.248
|
NM_001081419
|
Dip2a
|
DIP2 disco-interacting protein 2 homolog A (Drosophila)
|
|
chr8_+_14911653
|
3.214
|
NM_001037736
NM_172751
|
Arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr9_-_101101334
|
3.213
|
NM_172144
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr19_+_7568894
|
3.193
|
NM_146091
|
Atl3
|
atlastin GTPase 3
|
|
chr12_-_58647074
|
3.183
|
NM_008259
|
Foxa1
|
forkhead box A1
|
|
chr15_-_31297286
|
3.182
|
NM_001164441
NM_026153
NM_027496
|
Ankrd33b
|
ankyrin repeat domain 33B
|
|
chr3_-_107563372
|
3.127
|
NM_001113529
NM_001113530
NM_007778
|
Csf1
|
colony stimulating factor 1 (macrophage)
|
|
chr4_+_33069972
|
3.111
|
NM_027491
|
Rragd
|
Ras-related GTP binding D
|
|
chr11_-_59778306
|
3.064
|
NM_009026
|
Rasd1
|
RAS, dexamethasone-induced 1
|
|
chr2_+_113167862
|
3.059
|
NM_001043322
NM_010230
|
Fmn1
|
formin 1
|
|
chr12_+_85455277
|
3.059
|
NM_028821
|
Dnalc1
|
dynein, axonemal, light chain 1
|
|
chr13_+_64263131
|
3.040
|
NM_019986
|
Habp4
|
hyaluronic acid binding protein 4
|
|
chr11_+_75492764
|
3.033
|
NM_133656
|
Crk
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chr4_-_76240202
|
3.031
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr7_-_127038916
|
3.031
|
NM_173408
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr9_+_59598963
|
3.027
|
NM_173018
|
Myo9a
|
myosin IXa
|
|
chr8_-_86671651
|
2.979
|
NM_145970
|
Cc2d1a
|
coiled-coil and C2 domain containing 1A
|
|
chr6_+_122770201
|
2.976
|
NM_021899
|
Igfbp5
Foxj2
|
insulin-like growth factor binding protein 5
forkhead box J2
|
|
chr13_+_45485106
|
2.967
|
NM_153789
NM_181043
|
Mylip
|
myosin regulatory light chain interacting protein
|
|
chr10_+_94039007
|
2.910
|
NM_177026
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr6_-_90666474
|
2.902
|
NM_001134383
|
Iqsec1
|
IQ motif and Sec7 domain 1
|
|
chr1_+_59538966
|
2.901
|
NM_008057
|
Fzd7
|
frizzled homolog 7 (Drosophila)
|
|
chrX_-_159081510
|
2.894
|
NM_178256
|
Reps2
|
RALBP1 associated Eps domain containing protein 2
|
|
chr17_-_66868909
|
2.859
|
NM_024448
|
Rab12
|
RAB12, member RAS oncogene family
|
|
chr6_-_108135548
|
2.853
|
NM_145937
|
Sumf1
|
sulfatase modifying factor 1
|