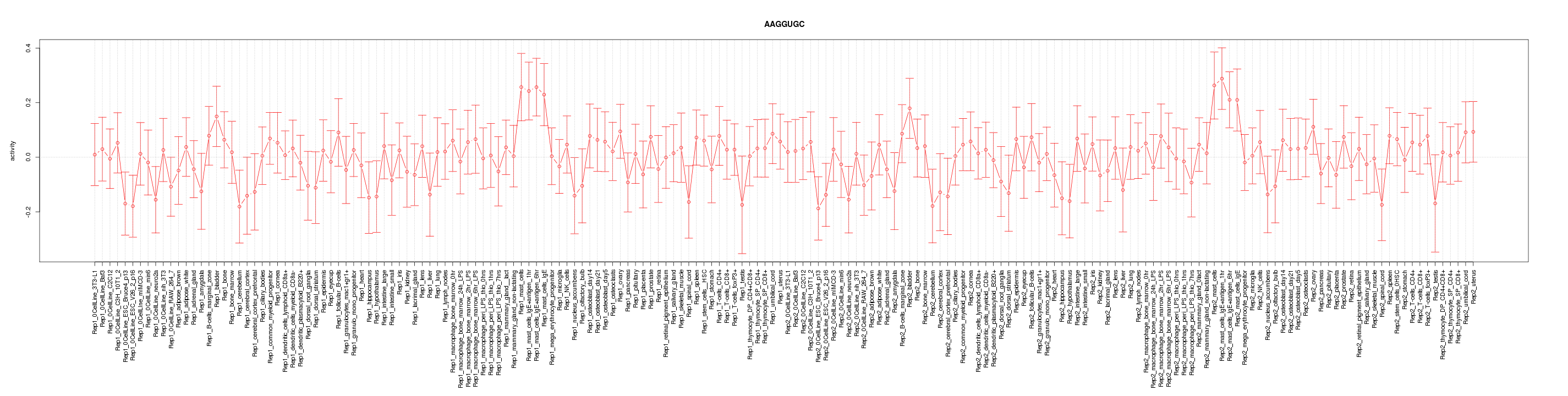
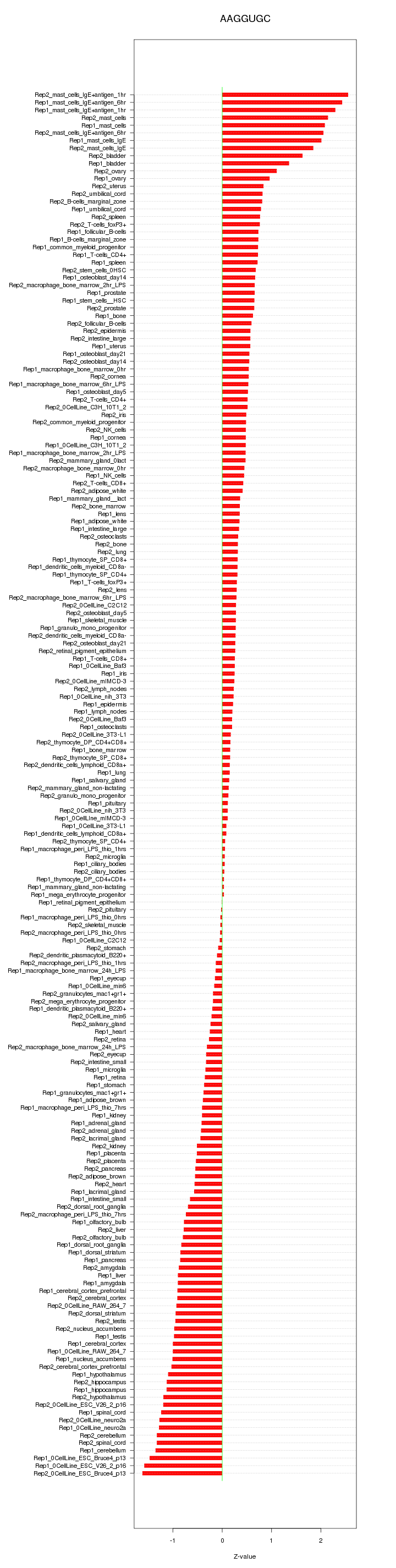
|
chr16_-_92697567
|
14.820
|
NM_001111023
NM_009821
|
Runx1
|
runt related transcription factor 1
|
|
chr16_-_92826307
|
11.456
|
NM_001111021
NM_001111022
|
Runx1
|
runt related transcription factor 1
|
|
chr3_+_60305108
|
10.912
|
NM_020007
|
Mbnl1
|
muscleblind-like 1 (Drosophila)
|
|
chr15_+_78813475
|
10.056
|
NM_001024716
|
Triobp
|
TRIO and F-actin binding protein
|
|
chr13_-_41454358
|
10.053
|
NM_017464
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9
|
|
chr10_-_18731766
|
9.991
|
NM_001166402
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr16_+_24721940
|
9.801
|
NM_001145954
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr5_+_75971011
|
9.279
|
NM_001122733
NM_021099
|
Kit
|
kit oncogene
|
|
chr16_+_24393435
|
9.014
|
NM_178665
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr16_+_24448176
|
8.644
|
NM_001145952
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_-_18735154
|
8.078
|
NM_009397
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr10_+_40069113
|
7.989
|
NM_001168304
NM_198164
|
Cdk19
|
cyclin-dependent kinase 19
|
|
chr5_+_97426665
|
7.796
|
NM_080708
|
Bmp2k
|
BMP2 inducible kinase
|
|
chr3_-_30868320
|
7.532
|
NM_001165954
NM_001165955
NM_001165956
NM_153421
|
Phc3
|
polyhomeotic-like 3 (Drosophila)
|
|
chr13_-_41582649
|
7.053
|
NM_001111324
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9
|
|
chr15_+_78778153
|
6.952
|
NM_001039155
NM_001039156
NM_138579
|
Triobp
|
TRIO and F-actin binding protein
|
|
chr2_+_112106470
|
6.856
|
NM_133649
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr8_-_36558514
|
6.053
|
NM_026067
|
Eri1
|
exoribonuclease 1
|
|
chr5_+_32438843
|
6.023
|
NM_008037
|
Fosl2
|
fos-like antigen 2
|
|
chr13_-_46060325
|
5.973
|
NM_009124
|
Atxn1
|
ataxin 1
|
|
chr4_+_118102305
|
5.570
|
NM_001039176
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
|
|
chr2_+_112124767
|
5.280
|
NM_133648
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr6_+_133985717
|
4.735
|
NM_007961
|
Etv6
|
ets variant gene 6 (TEL oncogene)
|
|
chr11_+_69579376
|
4.724
|
NM_029348
|
Zbtb4
|
zinc finger and BTB domain containing 4
|
|
chr11_-_94538693
|
4.722
|
NM_145828
|
Xylt2
|
xylosyltransferase II
|
|
chr15_+_80541675
|
4.572
|
NM_144812
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chr13_-_112599181
|
4.489
|
NM_011945
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr6_-_134742645
|
4.384
|
NM_001048054
NM_130447
|
Dusp16
|
dual specificity phosphatase 16
|
|
chr4_+_118100682
|
4.377
|
NM_001039175
NM_019422
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
|
|
chr7_+_134913327
|
4.119
|
NM_198424
|
Orai3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr9_+_30838224
|
4.116
|
NM_001115130
NM_172765
|
Zbtb44
|
zinc finger and BTB domain containing 44
|
|
chr6_+_140572056
|
4.105
|
NM_001005605
|
Aebp2
|
AE binding protein 2
|
|
chr18_-_39646734
|
4.048
|
NM_008173
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1
|
|
chr4_+_64785207
|
4.036
|
NM_021362
|
Pappa
|
pregnancy-associated plasma protein A
|
|
chr8_-_112261607
|
4.005
|
NM_001080930
|
Atxn1l
|
ataxin 1-like
|
|
chr16_-_50432421
|
3.903
|
NM_027444
|
Bbx
|
bobby sox homolog (Drosophila)
|
|
chr18_+_76401531
|
3.793
|
NM_010754
|
Smad2
|
MAD homolog 2 (Drosophila)
|
|
chr2_+_163899842
|
3.766
|
NM_021420
|
Stk4
|
serine/threonine kinase 4
|
|
chr10_+_43198945
|
3.651
|
NM_199028
|
Bend3
|
BEN domain containing 3
|
|
chr9_+_118835589
|
3.530
|
NM_133710
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr2_-_34769479
|
3.361
|
NM_028716
|
Phf19
|
PHD finger protein 19
|
|
chr1_+_136456754
|
3.361
|
NM_152895
|
Kdm5b
|
lysine (K)-specific demethylase 5B
|
|
chr10_-_5734448
|
3.248
|
NM_007956
|
Esr1
|
estrogen receptor 1 (alpha)
|
|
chr1_-_63160764
|
3.246
|
NM_001081436
NM_001114609
|
Ino80d
|
INO80 complex subunit D
|
|
chr5_-_73647856
|
3.239
|
NM_028194
|
Fryl
|
furry homolog-like (Drosophila)
|
|
chr12_+_75008853
|
3.202
|
NM_010431
|
Hif1a
|
hypoxia inducible factor 1, alpha subunit
|
|
chr11_+_4167080
|
3.129
|
NM_001039537
|
Lif
|
leukemia inhibitory factor
|
|
chr15_+_80629020
|
2.977
|
NM_177124
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chr19_+_32832002
|
2.921
|
NM_008960
|
Pten
|
phosphatase and tensin homolog
|
|
chr3_+_97817440
|
2.903
|
NM_010928
|
Notch2
|
Notch gene homolog 2 (Drosophila)
|
|
chr13_-_48966240
|
2.897
|
NM_011078
|
Phf2
|
PHD finger protein 2
|
|
chr11_+_4157570
|
2.818
|
NM_008501
|
Lif
|
leukemia inhibitory factor
|
|
chr3_-_100914081
|
2.520
|
NM_011197
|
Ptgfrn
|
prostaglandin F2 receptor negative regulator
|
|
chr10_+_87321789
|
2.473
|
NM_001111275
NM_010512
|
Igf1
|
insulin-like growth factor 1
|
|
chr2_-_24775083
|
2.369
|
NM_001012518
NM_001109686
NM_001109687
NM_172545
|
Ehmt1
|
euchromatic histone methyltransferase 1
|
|
chr16_+_94085504
|
2.313
|
NM_011377
|
Sim2
|
single-minded homolog 2 (Drosophila)
|
|
chr17_+_86020173
|
2.290
|
NM_011381
|
Six3
|
sine oculis-related homeobox 3 homolog (Drosophila)
|
|
chr17_+_78599556
|
2.185
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr7_+_108257288
|
2.141
|
NM_001146010
NM_199012
|
Fchsd2
|
FCH and double SH3 domains 2
|
|
chr16_+_44139908
|
2.099
|
NM_028108
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr6_+_140571183
|
2.096
|
NM_178803
|
Aebp2
|
AE binding protein 2
|
|
chr2_+_36085945
|
2.065
|
NM_008969
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1
|
|
chr3_-_9610062
|
1.973
|
NM_133218
|
Zfp704
|
zinc finger protein 704
|
|
chr8_+_47825098
|
1.906
|
NM_008391
|
Irf2
|
interferon regulatory factor 2
|
|
chr4_+_105988790
|
1.905
|
NM_183225
|
Usp24
|
ubiquitin specific peptidase 24
|
|
chr13_-_94269616
|
1.885
|
NM_021310
|
Jmy
|
junction-mediating and regulatory protein
|
|
chr1_+_40496493
|
1.881
|
NM_010743
|
Il1rl1
|
interleukin 1 receptor-like 1
|
|
chr12_+_3426883
|
1.855
|
NM_172421
|
Asxl2
|
additional sex combs like 2 (Drosophila)
|
|
chr11_+_77499638
|
1.750
|
NM_001024205
|
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr3_+_96500993
|
1.712
|
NM_001165949
NM_018812
|
Pias3
|
protein inhibitor of activated STAT 3
|
|
chr9_-_53344748
|
1.668
|
NM_007499
|
Atm
|
ataxia telangiectasia mutated homolog (human)
|
|
chr13_-_74429744
|
1.657
|
NM_009644
|
Ahrr
|
aryl-hydrocarbon receptor repressor
|
|
chr9_-_88376937
|
1.649
|
NM_019666
NM_019796
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr3_-_38383688
|
1.646
|
NM_001167883
|
Ankrd50
|
ankyrin repeat domain 50
|
|
chr10_+_79957650
|
1.579
|
NM_139226
|
Onecut3
|
one cut domain, family member 3
|
|
chr3_-_146475652
|
1.569
|
NM_011100
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr11_+_58028478
|
1.523
|
NM_019440
|
Irgm2
|
immunity-related GTPase family M member 2
|
|
chr8_-_70498472
|
1.462
|
NM_177698
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr14_-_20642973
|
1.392
|
NM_026528
|
2700060E02Rik
|
RIKEN cDNA 2700060E02 gene
|
|
chr12_-_85559793
|
1.383
|
NM_001163501
|
C130039O16Rik
|
RIKEN cDNA C130039O16 gene
|
|
chr13_+_34094742
|
1.350
|
NM_009068
|
Ripk1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1
|
|
chr2_+_72123693
|
1.276
|
NM_023057
NM_001164791
NM_178084
|
B230120H23Rik
|
RIKEN cDNA B230120H23 gene
|
|
chr6_+_108778633
|
1.273
|
NM_138677
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr1_-_132994093
|
1.234
|
NM_008551
|
Mapkapk2
|
MAP kinase-activated protein kinase 2
|
|
chr11_+_59120429
|
1.216
|
NM_139298
|
Wnt9a
|
wingless-type MMTV integration site 9A
|
|
chr5_-_116474254
|
1.206
|
NM_031869
|
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr2_+_117937623
|
1.163
|
NM_011580
|
Thbs1
|
thrombospondin 1
|
|
chr9_-_35007875
|
1.154
|
NM_001177845
|
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein
|
|
chr11_+_29273673
|
1.147
|
NM_176841
|
Ccdc88a
|
coiled coil domain containing 88A
|
|
chr16_-_85901362
|
1.134
|
NM_011782
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2)
|
|
chr16_-_22163264
|
1.117
|
NM_183029
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr14_+_70945429
|
1.109
|
NM_180588
|
Reep4
|
receptor accessory protein 4
|
|
chr18_+_66618186
|
1.025
|
NM_021451
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr2_+_154959216
|
1.010
|
NM_008395
|
Itch
|
itchy, E3 ubiquitin protein ligase
|
|
chr15_+_73578455
|
1.005
|
NM_008975
|
Ptp4a3
|
protein tyrosine phosphatase 4a3
|
|
chr13_-_64254466
|
0.984
|
NM_175494
|
Zfp367
|
zinc finger protein 367
|
|
chr2_-_153067043
|
0.983
|
NM_018807
|
Plagl2
|
pleiomorphic adenoma gene-like 2
|
|
chr9_-_35007458
|
0.974
|
NM_001177847
NM_001177846
NM_054096
|
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein
|
|
chr16_-_44028057
|
0.952
|
NM_001172107
|
Gramd1c
|
GRAM domain containing 1C
|
|
chr2_+_153067186
|
0.951
|
NM_080463
|
Pofut1
|
protein O-fucosyltransferase 1
|
|
chr15_+_10882086
|
0.942
|
NM_030888
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3
|
|
chr3_+_96500294
|
0.928
|
NM_146135
|
Pias3
|
protein inhibitor of activated STAT 3
|
|
chr1_+_52176281
|
0.924
|
NM_009283
|
Stat1
|
signal transducer and activator of transcription 1
|
|
chr6_+_86992796
|
0.909
|
NM_013528
|
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1
|
|
chr10_-_127930880
|
0.905
|
NM_010860
|
Myl6
|
myosin, light polypeptide 6, alkali, smooth muscle and non-muscle
|
|
chr16_+_32332294
|
0.887
|
NM_177633
|
Ubxn7
|
UBX domain protein 7
|
|
chr12_-_92828018
|
0.878
|
NM_031391
|
Gtf2a1
|
general transcription factor II A, 1
|
|
chr3_+_88489708
|
0.871
|
NM_028814
|
2810403A07Rik
|
RIKEN cDNA 2810403A07 gene
|
|
chr4_+_147314951
|
0.852
|
NM_029841
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene
|
|
chr16_+_34785006
|
0.845
|
NM_139300
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr7_+_17401237
|
0.840
|
NM_001039878
NM_133789
|
Strn4
|
striatin, calmodulin binding protein 4
|
|
chrX_+_102275071
|
0.833
|
NM_026312
|
2610029G23Rik
|
RIKEN cDNA 2610029G23 gene
|
|
chr5_+_139856534
|
0.819
|
NM_030258
|
Gpr146
|
G protein-coupled receptor 146
|
|
chr16_-_44016471
|
0.811
|
NM_153528
|
Gramd1c
|
GRAM domain containing 1C
|
|
chr5_+_91503642
|
0.805
|
NM_007950
|
Ereg
|
epiregulin
|
|
chr17_-_30024821
|
0.776
|
NM_001081160
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr19_+_53218486
|
0.769
|
NM_013758
|
Add3
|
adducin 3 (gamma)
|
|
chr8_+_109817355
|
0.767
|
NM_018823
NM_133957
|
Nfat5
|
nuclear factor of activated T-cells 5
|
|
chr8_+_72315061
|
0.761
|
NM_133224
|
Atp13a1
|
ATPase type 13A1
|
|
chr1_-_45560001
|
0.751
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr6_+_55286292
|
0.745
|
NM_007472
|
Aqp1
|
aquaporin 1
|
|
chr2_-_52601140
|
0.727
|
NM_019667
|
Stam2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr3_+_14641726
|
0.723
|
NM_024495
|
Car13
|
carbonic anhydrase 13
|
|
chr19_+_57435498
|
0.711
|
NM_145505
|
Fam160b1
|
family with sequence similarity 160, member B1
|
|
chrX_-_16394443
|
0.699
|
NM_172778
|
Maob
|
monoamine oxidase B
|
|
chr10_-_128082881
|
0.676
|
NM_011772
|
Ikzf4
|
IKAROS family zinc finger 4
|
|
chr12_-_85534964
|
0.655
|
NM_001163502
|
C130039O16Rik
|
RIKEN cDNA C130039O16 gene
|
|
chr10_+_87323855
|
0.625
|
NM_001111274
NM_001111276
NM_184052
|
Igf1
|
insulin-like growth factor 1
|
|
chr19_+_21346734
|
0.624
|
NM_009551
|
Zfand5
|
zinc finger, AN1-type domain 5
|
|
chr10_+_68675404
|
0.609
|
NM_001081347
|
Rhobtb1
|
Rho-related BTB domain containing 1
|
|
chr11_+_86298106
|
0.597
|
NM_029788
|
Rnft1
|
ring finger protein, transmembrane 1
|
|
chr5_+_139865857
|
0.596
|
NM_001038703
|
Gpr146
|
G protein-coupled receptor 146
|
|
chr5_+_130695613
|
0.589
|
NM_001081394
|
0610007L01Rik
|
RIKEN cDNA 0610007L01 gene
|
|
chr18_+_53624169
|
0.577
|
NM_001033281
|
Prdm6
|
PR domain containing 6
|
|
chr17_-_32925218
|
0.570
|
NM_172458
|
Zfp871
|
zinc finger protein 871
|
|
chr14_-_32064403
|
0.554
|
NM_027949
|
Phf7
|
PHD finger protein 7
|
|
chr13_-_104124527
|
0.547
|
NM_175171
|
Mast4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr18_+_9957913
|
0.533
|
NM_153552
|
Thoc1
|
THO complex 1
|
|
chr11_+_49016828
|
0.532
|
NM_001024846
NM_009562
|
Zfp62
|
zinc finger protein 62
|
|
chr17_+_64287320
|
0.523
|
NM_008000
|
Fert2
|
fer (fms/fps related) protein kinase, testis specific 2
|
|
chr18_-_78320126
|
0.522
|
NM_001171010
NM_028122
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1
|
|
chr18_-_78338857
|
0.516
|
NM_001171011
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1
|
|
chr5_+_136514365
|
0.513
|
NM_175309
|
Upk3b
|
uroplakin 3B
|
|
chr4_-_15940946
|
0.502
|
NM_145950
|
Osgin2
|
oxidative stress induced growth inhibitor family member 2
|
|
chr18_+_32227377
|
0.501
|
NM_173441
|
Iws1
|
IWS1 homolog (S. cerevisiae)
|
|
chr6_+_21899614
|
0.494
|
NM_023626
|
Ing3
|
inhibitor of growth family, member 3
|
|
chr1_+_70772288
|
0.492
|
NM_177164
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like
|
|
chr8_-_48760457
|
0.482
|
NM_023503
|
Ing2
|
inhibitor of growth family, member 2
|
|
chr1_-_137481914
|
0.467
|
NM_173437
|
Nav1
|
neuron navigator 1
|
|
chr6_+_134961629
|
0.460
|
NM_026360
|
Ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47
|
|
chr17_+_53706283
|
0.458
|
NM_020005
|
Kat2b
|
K(lysine) acetyltransferase 2B
|
|
chr17_+_26852581
|
0.456
|
NM_029870
|
A930001N09Rik
|
RIKEN cDNA A930001N09 gene
|
|
chr1_+_19198994
|
0.446
|
NM_009334
|
Tcfap2b
|
transcription factor AP-2 beta
|
|
chr11_-_77421308
|
0.443
|
NM_144825
|
Taok1
|
TAO kinase 1
|
|
chrX_-_141921588
|
0.442
|
NM_153319
|
Amot
|
angiomotin
|
|
chr5_-_137548123
|
0.437
|
NM_008871
|
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1
|
|
chr4_-_155366696
|
0.419
|
NM_080445
|
B3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase, polypeptide 6
|
|
chr6_+_86354212
|
0.417
|
NM_001164078
NM_001164079
NM_011585
|
Tia1
|
cytotoxic granule-associated RNA binding protein 1
|
|
chr5_+_73039034
|
0.414
|
NM_001081205
|
Nipal1
|
NIPA-like domain containing 1
|
|
chr17_-_56323312
|
0.414
|
NM_080837
|
D17Wsu104e
|
DNA segment, Chr 17, Wayne State University 104, expressed
|
|
chr1_-_12980994
|
0.413
|
NM_172841
|
Slco5a1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr1_-_162964371
|
0.411
|
NM_173424
|
Zbtb37
|
zinc finger and BTB domain containing 37
|
|
chr2_-_56967331
|
0.404
|
NM_013613
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr6_-_119988624
|
0.401
|
NM_001185020
NM_001185021
NM_198703
|
Wnk1
|
WNK lysine deficient protein kinase 1
|
|
chr15_+_80453912
|
0.401
|
NM_010815
|
Grap2
|
GRB2-related adaptor protein 2
|
|
chr17_+_53723450
|
0.389
|
NM_001190846
|
Kat2b
|
K(lysine) acetyltransferase 2B
|
|
chr17_+_64245366
|
0.386
|
NM_001037997
|
Fert2
|
fer (fms/fps related) protein kinase, testis specific 2
|
|
chr15_-_56526052
|
0.379
|
NM_008216
|
Has2
|
hyaluronan synthase 2
|
|
chr11_+_51398258
|
0.367
|
NM_028398
|
Agxt2l2
|
alanine-glyoxylate aminotransferase 2-like 2
|
|
chr17_-_66300454
|
0.353
|
NM_023053
|
Twsg1
|
twisted gastrulation homolog 1 (Drosophila)
|
|
chr19_+_53214934
|
0.341
|
NM_001164099
NM_001164101
|
Add3
|
adducin 3 (gamma)
|
|
chr8_-_120195414
|
0.339
|
NM_028725
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr19_-_42687234
|
0.333
|
NM_001164311
NM_053083
|
Loxl4
|
lysyl oxidase-like 4
|
|
chr15_+_73555336
|
0.327
|
NM_001166388
|
Ptp4a3
|
protein tyrosine phosphatase 4a3
|
|
chr15_+_99869071
|
0.321
|
NM_001159361
|
Dip2b
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr1_-_43155380
|
0.320
|
NM_001013025
|
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1
|
|
chr9_-_14185685
|
0.304
|
NM_028013
|
Endod1
|
endonuclease domain containing 1
|
|
chr18_+_62483857
|
0.304
|
NM_008313
|
Htr4
|
5 hydroxytryptamine (serotonin) receptor 4
|
|
chr3_+_54559503
|
0.299
|
NM_019483
|
Smad9
|
MAD homolog 9 (Drosophila)
|
|
chr4_+_114079291
|
0.291
|
NM_001085549
|
Gm12824
|
predicted gene 12824
|
|
chr9_-_65732843
|
0.289
|
NM_001170907
|
Trip4
|
thyroid hormone receptor interactor 4
|
|
chr1_+_82583495
|
0.286
|
NM_007734
|
Col4a3
|
collagen, type IV, alpha 3
|
|
chr10_+_57206184
|
0.284
|
NM_008297
|
Hsf2
|
heat shock factor 2
|
|
chr3_-_146433200
|
0.280
|
NM_001164198
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr14_+_65977510
|
0.280
|
NM_199029
|
Zfp395
|
zinc finger protein 395
|
|
chr6_+_34813131
|
0.267
|
NM_197986
|
Tmem140
|
transmembrane protein 140
|
|
chr5_-_139060952
|
0.263
|
NM_001044747
NM_001163797
NM_013844
|
Zfp68
|
zinc finger protein 68
|
|
chr1_-_33726582
|
0.258
|
NM_008922
|
Prim2
|
DNA primase, p58 subunit
|
|
chr6_-_39675456
|
0.254
|
NM_139294
|
Braf
|
Braf transforming gene
|
|
chr9_+_115818572
|
0.253
|
NM_028638
|
Gadl1
|
glutamate decarboxylase-like 1
|
|
chr15_+_73553574
|
0.249
|
NM_001166389
NM_001166390
|
Ptp4a3
|
protein tyrosine phosphatase 4a3
|
|
chr9_+_118387306
|
0.246
|
NM_001164789
NM_010136
|
Eomes
|
eomesodermin homolog (Xenopus laevis)
|
|
chr13_-_93914519
|
0.245
|
NM_023821
|
Cmya5
|
cardiomyopathy associated 5
|
|
chr4_-_139388882
|
0.245
|
NM_011039
|
Pax7
|
paired box gene 7
|
|
chr13_+_41701584
|
0.244
|
NM_001163572
|
Tmem170b
|
transmembrane protein 170B
|
|
chr2_+_91493364
|
0.241
|
NM_146124
|
Arhgap1
|
Rho GTPase activating protein 1
|
|
chr2_+_75497282
|
0.237
|
NM_053263
NM_146130
NM_198090
|
Hnrnpa3
Gm6793
|
heterogeneous nuclear ribonucleoprotein A3
heterogeneous nuclear ribonucleoprotein A3 pseudogene
|
|
chr7_-_91234908
|
0.233
|
NM_030728
|
9930013L23Rik
|
RIKEN cDNA 9930013L23 gene
|
|
chr5_+_137443701
|
0.231
|
NM_021719
|
Cldn15
|
claudin 15
|
|
chr1_+_19202046
|
0.227
|
NM_001025305
|
Tcfap2b
|
transcription factor AP-2 beta
|