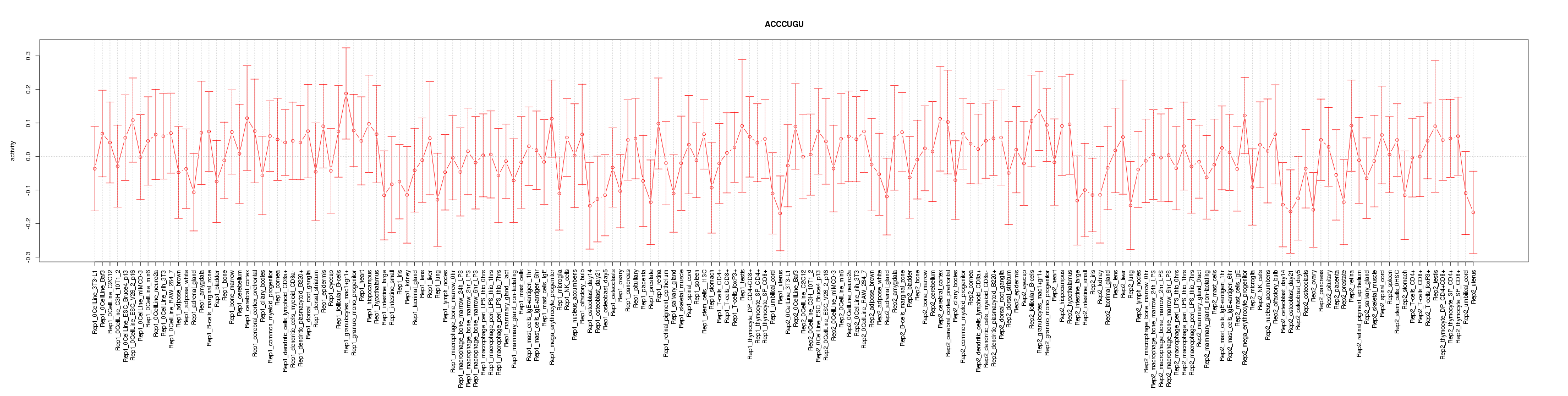
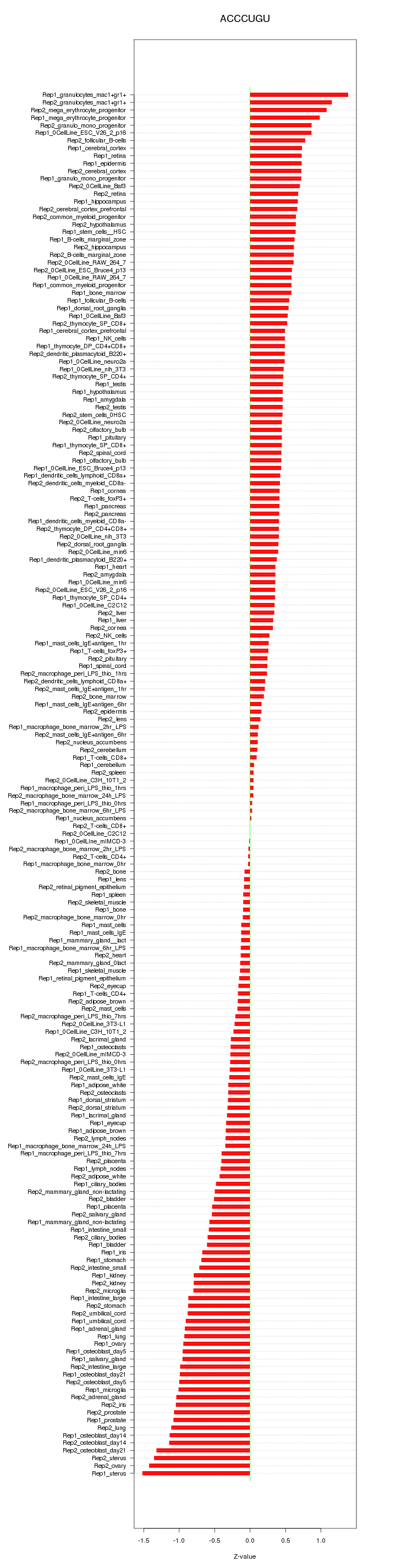
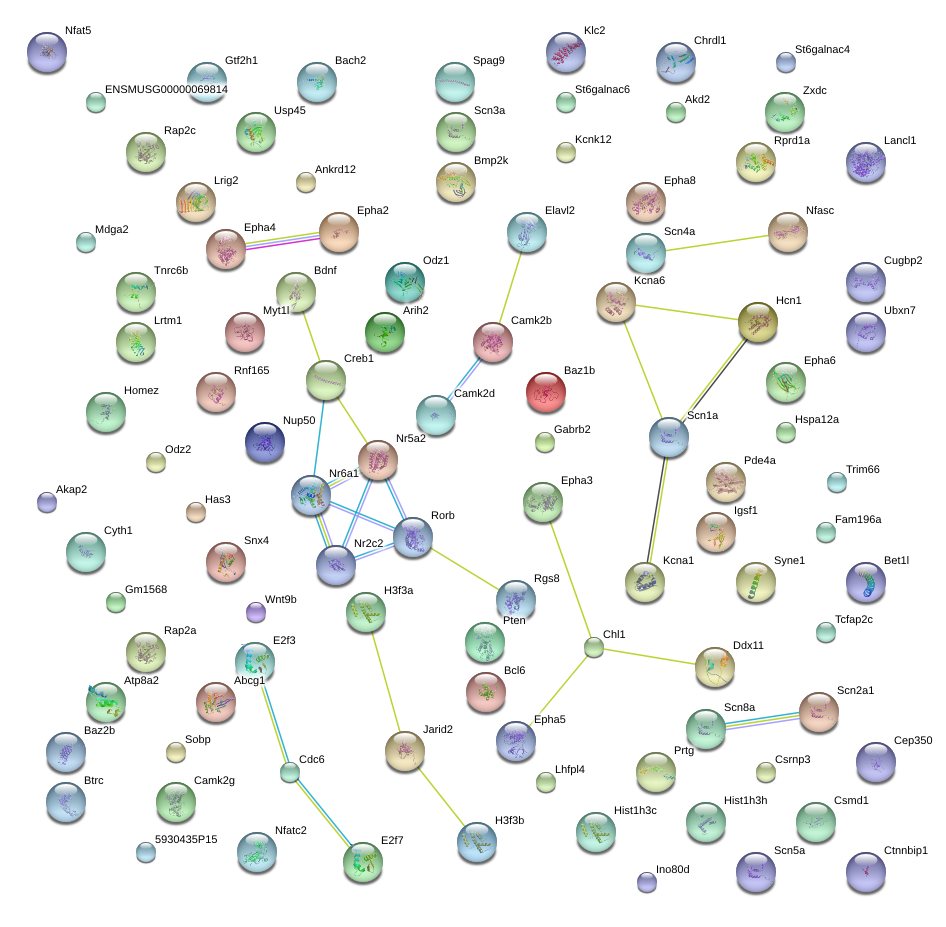
|
chr4_-_91066674
|
7.339
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_-_91042997
|
6.897
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_-_91038729
|
6.793
|
NM_010486
NM_207686
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_+_155500166
|
4.980
|
NM_026380
|
Rgs8
|
regulator of G-protein signaling 8
|
|
chr12_+_30219769
|
4.675
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr11_-_36757743
|
4.457
|
NM_011856
|
Odz2
|
odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr12_+_30213224
|
4.299
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr11_+_42233258
|
4.248
|
NM_008070
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2
|
|
chr11_+_98769464
|
4.214
|
NM_011799
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr11_+_98769162
|
4.113
|
NM_001025779
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr13_+_118391126
|
3.683
|
NM_010408
|
Hcn1
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 1
|
|
chr2_+_65683823
|
3.407
|
NM_153409
NM_178634
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3
|
|
chr1_-_134603848
|
3.246
|
NM_001160318
|
Nfasc
|
neurofascin
|
|
chr4_+_32504409
|
3.167
|
NM_001109661
|
Bach2
|
BTB and CNC homology 2
|
|
chr1_-_134539434
|
3.118
|
NM_001160316
|
Nfasc
|
neurofascin
|
|
chr12_-_68323528
|
2.977
|
NM_001193266
NM_207010
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr9_+_72655019
|
2.754
|
NM_175485
|
Prtg
|
protogenin homolog (Gallus gallus)
|
|
chr6_-_113145321
|
2.746
|
NM_177763
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4
|
|
chr9_+_21001148
|
2.733
|
NM_019798
|
Pde4a
|
phosphodiesterase 4A, cAMP specific
|
|
chr10_+_4795808
|
2.586
|
NM_153399
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr5_-_84846348
|
2.578
|
NM_007937
|
Epha5
|
Eph receptor A5
|
|
chr6_+_103460834
|
2.514
|
NM_007697
|
Chl1
|
cell adhesion molecule with homology to L1CAM
|
|
chr10_-_42894259
|
2.458
|
NM_175407
|
Sobp
|
sine oculis-binding protein homolog (Drosophila)
|
|
chr11_-_5965743
|
2.397
|
NM_001174053
NM_001174054
NM_007595
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr1_-_134638361
|
2.329
|
NM_001160317
NM_182716
|
Nfasc
|
neurofascin
|
|
chr7_-_116651647
|
2.268
|
NM_001170912
NM_181853
|
Trim66
|
tripartite motif-containing 66
|
|
chr7_-_116637251
|
2.222
|
NM_001170913
|
Trim66
|
tripartite motif-containing 66
|
|
chr18_-_24688688
|
1.957
|
NM_144861
|
Rprd1a
|
regulation of nuclear pre-mRNA domain containing 1A
|
|
chr4_+_57580989
|
1.918
|
NM_172868
|
Palm2
|
paralemmin 2
|
|
chr8_-_17535279
|
1.895
|
NM_053171
|
Csmd1
|
CUB and Sushi multiple domains 1
|
|
chr1_-_63160764
|
1.857
|
NM_001081436
NM_001114609
|
Ino80d
|
INO80 complex subunit D
|
|
chr18_-_77803843
|
1.797
|
NM_001164504
|
8030462N17Rik
Rnf165
|
RIKEN cDNA 8030462N17 gene
ring finger protein 165
|
|
chr14_-_55482884
|
1.789
|
NM_001177705
|
Homez
|
homeodomain leucine zipper-encoding gene
|
|
chr6_-_120443824
|
1.777
|
NM_033567
|
Cecr6
|
cat eye syndrome chromosome region, candidate 6 homolog (human)
|
|
chr11_-_118074052
|
1.761
|
NM_001112700
|
Cyth1
|
cytohesin 1
|
|
chr5_+_97426665
|
1.727
|
NM_080708
|
Bmp2k
|
BMP2 inducible kinase
|
|
chr9_+_20970150
|
1.725
|
NM_183408
|
Pde4a
|
phosphodiesterase 4A, cAMP specific
|
|
chr17_+_31194623
|
1.627
|
NM_009593
|
Abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr10_+_110182506
|
1.622
|
NM_178609
|
E2f7
|
E2F transcription factor 7
|
|
chr15_+_80629020
|
1.621
|
NM_177124
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chr13_+_41701584
|
1.617
|
NM_001163572
|
Tmem170b
|
transmembrane protein 170B
|
|
chr19_-_5118229
|
1.617
|
NM_008451
|
Klc2
|
kinesin light chain 2
|
|
chr12_-_81861527
|
1.591
|
NM_001008423
|
Gm1568
|
predicted gene 1568
|
|
chr19_-_58935470
|
1.569
|
NM_175199
|
Hspa12a
|
heat shock protein 12A
|
|
chr14_-_55479326
|
1.569
|
NM_183174
|
Homez
|
homeodomain leucine zipper-encoding gene
|
|
chr19_+_32832002
|
1.566
|
NM_008960
|
Pten
|
phosphatase and tensin homolog
|
|
chr8_+_109394141
|
1.553
|
NM_008217
|
Has3
|
hyaluronan synthase 3
|
|
chr11_-_103611082
|
1.517
|
NM_011719
|
Wnt9b
|
wingless-type MMTV integration site 9B
|
|
chr2_+_172375092
|
1.508
|
NM_009335
|
Tcfap2c
|
transcription factor AP-2, gamma
|
|
chr11_-_118109789
|
1.442
|
NM_001112699
NM_011180
|
Cyth1
|
cytohesin 1
|
|
chr15_+_80541675
|
1.400
|
NM_144812
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chr1_+_64579358
|
1.334
|
NM_001037726
NM_009952
NM_133828
|
Creb1
|
cAMP responsive element binding protein 1
|
|
chr2_-_7317824
|
1.233
|
NM_001110231
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_67085404
|
1.216
|
NM_001190984
NM_001190985
NM_021295
|
Lancl1
|
LanC (bacterial lantibiotic synthetase component C)-like 1
|
|
chr6_-_126690651
|
1.139
|
NM_013568
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6
|
|
chr11_+_74433092
|
1.138
|
NM_001013784
|
E130309D14Rik
|
RIKEN cDNA E130309D14 gene
|
|
chr19_+_45438212
|
1.134
|
NM_001037758
NM_009771
|
Btrc
|
beta-transducin repeat containing protein
|
|
chr2_+_172376490
|
1.086
|
NM_001159696
|
Tcfap2c
|
transcription factor AP-2, gamma
|
|
chr19_-_19075535
|
1.082
|
NM_146095
|
Rorb
|
RAR-related orphan receptor beta
|
|
chr8_+_109817355
|
1.027
|
NM_018823
NM_133957
|
Nfat5
|
nuclear factor of activated T-cells 5
|
|
chr17_-_66426361
|
0.986
|
NM_001025572
|
Ankrd12
|
ankyrin repeat domain 12
|
|
chr19_-_19185685
|
0.977
|
NM_001043354
|
Rorb
|
RAR-related orphan receptor beta
|
|
chr2_-_65405546
|
0.957
|
NM_018732
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha
|
|
chr7_-_135883942
|
0.847
|
NM_145955
|
Mcmbp
|
MCM (minichromosome maintenance deficient) binding protein
|
|
chr11_-_115885714
|
0.837
|
NM_008211
|
H3f3b
|
H3 histone, family 3B
|
|
chr4_-_136512727
|
0.798
|
NM_007939
|
Epha8
|
Eph receptor A8
|
|
chr9_-_108551631
|
0.794
|
NM_011790
|
Arih2
|
ariadne homolog 2 (Drosophila)
|
|
chr2_-_38640034
|
0.793
|
NM_001159548
NM_001159549
|
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr6_+_92041408
|
0.776
|
NM_011630
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr2_-_6805937
|
0.727
|
NM_001110228
NM_001160293
NM_010160
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_109514856
|
0.694
|
NM_007540
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr1_-_157820378
|
0.636
|
NM_001039184
|
Cep350
|
centrosomal protein 350
|
|
chr7_-_142129892
|
0.609
|
NM_001143802
|
Fam196a
|
family with sequence similarity 196, member A
|
|
chrX_-_47150892
|
0.593
|
NM_177591
NM_177915
|
Igsf1
|
immunoglobulin superfamily, member 1
|
|
chr16_+_32332294
|
0.559
|
NM_177633
|
Ubxn7
|
UBX domain protein 7
|
|
chr3_-_104315778
|
0.539
|
NM_001025067
|
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2
|
|
chr6_+_90319485
|
0.539
|
NM_030260
NM_173002
|
Zxdc
|
ZXD family zinc finger C
|
|
chr4_+_21703416
|
0.530
|
NM_152825
|
Usp45
|
ubiquitin specific petidase 45
|
|
chr15_+_84753817
|
0.524
|
NM_016714
|
Nup50
|
nucleoporin 50
|
|
chr2_-_59963796
|
0.516
|
NM_001001182
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr11_+_93857338
|
0.502
|
NM_027569
|
Spag9
|
sperm associated antigen 9
|
|
chr14_+_120877548
|
0.491
|
NM_029519
|
Rap2a
|
RAS related protein 2a
|
|
chr4_+_148919218
|
0.449
|
NM_001141930
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chr7_+_54051432
|
0.424
|
NM_008186
|
Gtf2h1
|
general transcription factor II H, polypeptide 1
|
|
chr6_-_126595818
|
0.419
|
NM_010595
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1
|
|
chr2_+_109532592
|
0.418
|
NM_001048141
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr13_+_44826121
|
0.413
|
NM_021878
|
Jarid2
|
jumonji, AT rich interactive domain 2
|
|
chr14_-_21613162
|
0.385
|
NM_001039138
NM_001039139
NM_178597
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr4_+_148892231
|
0.383
|
NM_023465
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chr16_-_23988680
|
0.333
|
NM_009744
|
Bcl6
|
B-cell leukemia/lymphoma 6
|
|
chr2_-_168427037
|
0.320
|
NM_001037178
|
Nfatc2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr14_+_49065971
|
0.315
|
NM_172600
|
6720456H20Rik
|
RIKEN cDNA 6720456H20 gene
|
|
chr5_+_135663096
|
0.293
|
NM_011714
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr14_+_29831283
|
0.273
|
NM_176920
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1
|
|
chr2_+_32462471
|
0.228
|
NM_001025310
NM_016973
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr2_+_32455168
|
0.222
|
NM_001025311
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr1_-_138856866
|
0.219
|
NM_030676
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr2_+_109533719
|
0.168
|
NM_001048142
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr16_+_33251485
|
0.109
|
NM_080557
|
Snx4
|
sorting nexin 4
|
|
chr13_-_30077931
|
0.079
|
NM_010093
|
E2f3
|
E2F transcription factor 3
|
|
chr5_-_125659422
|
0.064
|
NM_011424
|
Ncor2
|
nuclear receptor co-repressor 2
|
|
chr4_-_132478424
|
0.064
|
NM_001163792
NM_145553
|
Fam76a
|
family with sequence similarity 76, member A
|
|
chr11_+_79153388
|
0.043
|
NM_010897
|
Nf1
|
neurofibromatosis 1
|
|
chr5_+_120284671
|
0.022
|
NM_011537
|
Tbx5
|
T-box 5
|
|
chr1_-_138850206
|
0.013
|
NM_001159769
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr11_+_72503487
|
0.011
|
NM_009671
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1
|