|
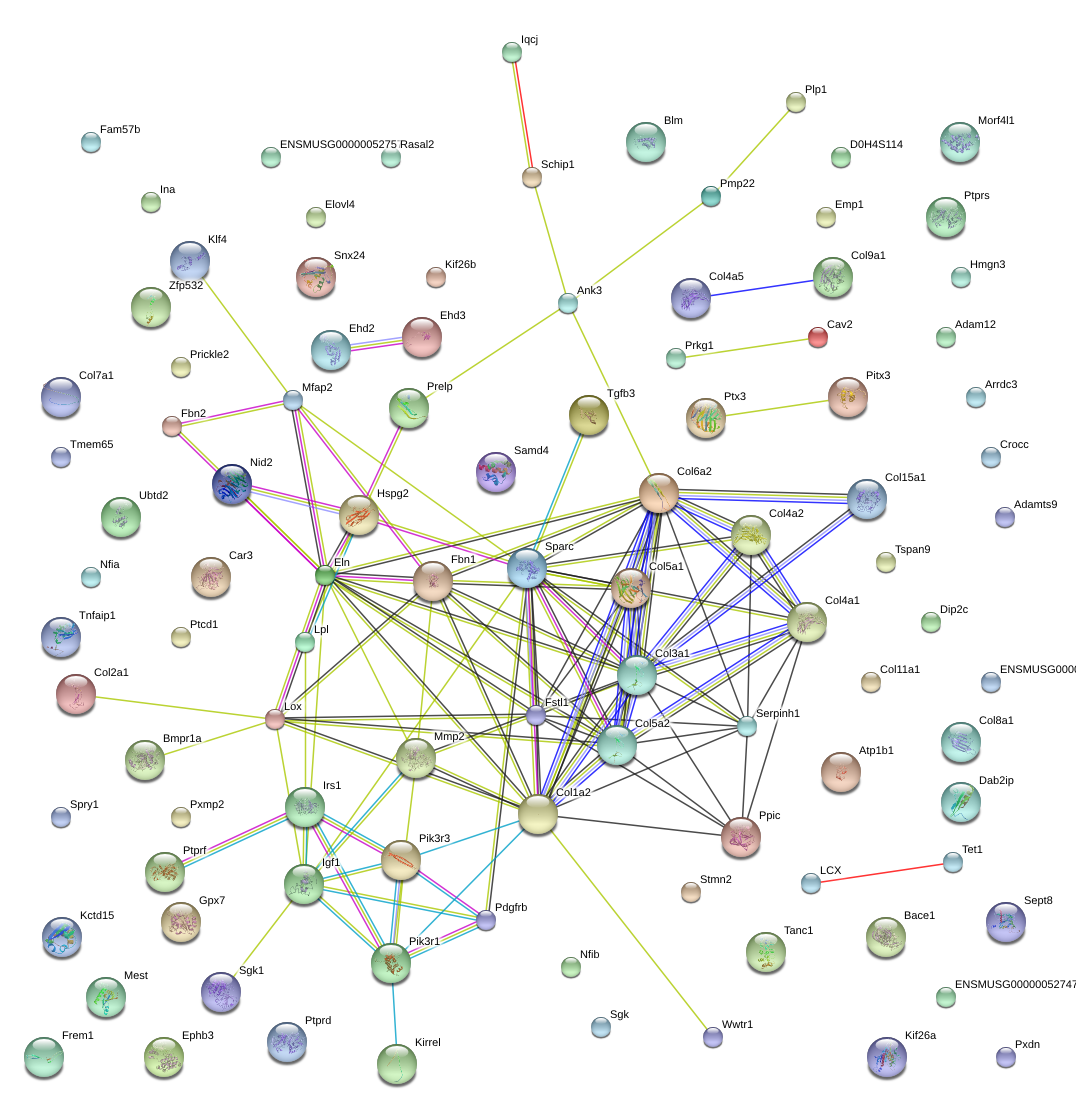
chr11_-_55233581
|
60.347
|
NM_009242
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr16_+_37777075
|
50.340
|
NM_008047
|
Fstl1
|
follistatin-like 1
|
|
chr18_-_33623668
|
46.023
|
NM_001109988
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr18_-_33623400
|
45.491
|
NM_001109989
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr18_-_33623069
|
44.678
|
NM_001109990
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr18_-_33623287
|
44.221
|
NM_053078
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr6_+_4455686
|
43.404
|
NM_007743
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr8_-_11312729
|
40.403
|
NM_009931
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr7_-_106501642
|
40.205
|
NM_001111043
NM_001111044
NM_009825
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1
|
|
chr8_+_95351212
|
39.759
|
NM_008610
|
Mmp2
|
matrix metallopeptidase 2
|
|
chr8_+_11312828
|
37.407
|
NM_009932
|
Col4a2
|
collagen, type IV, alpha 2
|
|
chr1_+_45368382
|
35.292
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_-_45560001
|
35.080
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr12_+_30622876
|
33.854
|
NM_181395
|
Pxdn
|
peroxidasin homolog (Drosophila)
|
|
chr6_+_30688050
|
33.697
|
NM_008590
|
Mest
|
mesoderm specific transcript
|
|
chr3_+_67868723
|
30.150
|
NM_001113420
NM_001113421
|
Schip1
|
schwannomin interacting protein 1
|
|
chr3_+_113733457
|
29.917
|
NM_007729
|
Col11a1
|
collagen, type XI, alpha 1
|
|
chr4_+_140566338
|
27.357
|
NM_001161799
|
Mfap2
|
microfibrillar-associated protein 2
|
|
chr18_-_53577625
|
27.286
|
NM_008908
|
Ppic
|
peptidylprolyl isomerase C
|
|
chr4_+_115894518
|
26.583
|
NM_181585
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55)
|
|
chr6_-_92431385
|
26.385
|
NM_001134460
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr4_-_108079288
|
26.349
|
NM_024198
|
Gpx7
|
glutathione peroxidase 7
|
|
chr12_-_87419813
|
26.178
|
NM_009368
|
Tgfb3
|
transforming growth factor, beta 3
|
|
chr6_-_92656148
|
26.062
|
NM_001081146
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr6_-_92517359
|
25.821
|
NM_001134461
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr2_-_125332107
|
25.485
|
NM_007993
|
Fbn1
|
fibrillin 1
|
|
chr4_+_140566551
|
25.237
|
NM_008546
|
Mfap2
|
microfibrillar-associated protein 2
|
|
chr3_+_68376289
|
25.079
|
NM_013928
|
Schip1
|
schwannomin interacting protein 1
|
|
chr6_-_92484838
|
24.887
|
NM_001134459
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr1_-_82287990
|
23.465
|
NM_010570
|
Irs1
|
insulin receptor substrate 1
|
|
chr18_+_61204798
|
23.227
|
NM_001146268
NM_008809
|
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide
|
|
chr18_+_65739883
|
23.126
|
NM_207255
|
Zfp532
|
zinc finger protein 532
|
|
chr3_-_57379831
|
22.921
|
NM_001168281
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr10_-_76086144
|
22.486
|
NM_146007
|
Col6a2
|
collagen, type VI, alpha 2
|
|
chr2_+_27741864
|
22.467
|
NM_015734
|
Col5a1
|
collagen, type V, alpha 1
|
|
chr16_-_57754816
|
22.462
|
NM_007739
|
Col8a1
|
collagen, type VIII, alpha 1
|
|
chr3_+_8509479
|
20.748
|
NM_025285
|
Stmn2
|
stathmin-like 2
|
|
chr14_+_20570444
|
20.719
|
NM_008695
|
Nid2
|
nidogen 2
|
|
chr3_-_57379600
|
20.545
|
NM_133784
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr7_-_35437859
|
20.435
|
NM_146188
|
Kctd15
|
potassium channel tetramerisation domain containing 15
|
|
chr1_+_180459245
|
20.288
|
NM_001161665
|
Kif26b
|
kinesin family member 26B
|
|
chr11_+_62944973
|
20.125
|
NM_008885
|
Pmp22
|
peripheral myelin protein 22
|
|
chr1_-_135817945
|
19.237
|
NM_054077
|
Prelp
|
proline arginine-rich end leucine-rich repeat
|
|
chr2_+_35477498
|
18.579
|
NM_001114124
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr2_+_59450080
|
18.451
|
NM_198294
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr10_+_69388242
|
18.081
|
NM_009670
NM_170687
|
Ank3
|
ankyrin 3, epithelial
|
|
chr3_+_66023808
|
17.942
|
NM_008987
|
Ptx3
|
pentraxin related gene
|
|
chr2_+_35547418
|
17.643
|
NM_001001602
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr10_+_68996455
|
17.613
|
NM_146005
NM_170688
NM_170689
NM_170690
NM_170728
NM_170729
NM_170730
|
Ank3
|
ankyrin 3, epithelial
|
|
chr1_-_166388431
|
17.524
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr3_+_37538808
|
17.452
|
NM_011896
|
Spry1
|
sprouty homolog 1 (Drosophila)
|
|
chrX_+_137909951
|
17.121
|
NM_001163155
NM_007736
|
Col4a5
|
collagen, type IV, alpha 5
|
|
chr4_-_75779379
|
16.765
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr9_+_45646611
|
16.715
|
NM_001145947
NM_011792
|
Bace1
|
beta-site APP cleaving enzyme 1
|
|
chr1_-_159342713
|
16.572
|
NM_177644
|
Rasal2
|
RAS protein activator like 2
|
|
chr14_+_47621014
|
16.147
|
NM_001163433
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr9_-_83040061
|
15.974
|
NM_026122
NM_175074
|
Hmgn3
|
high mobility group nucleosomal binding domain 3
|
|
chr11_+_53333270
|
15.957
|
NM_033144
|
Sept8
|
septin 8
|
|
chr4_-_55545337
|
15.931
|
NM_010637
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr14_-_35315715
|
15.874
|
NM_009758
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A
|
|
chr10_+_87321789
|
15.597
|
NM_001111275
NM_010512
|
Igf1
|
insulin-like growth factor 1
|
|
chr15_-_58654964
|
15.458
|
NM_175212
|
Tmem65
|
transmembrane protein 65
|
|
chr4_-_76240202
|
14.860
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_+_21714471
|
14.850
|
NM_011361
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_92851434
|
14.753
|
NM_175314
|
Adamts9
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9
|
|
chr10_+_21712008
|
14.614
|
NM_001161849
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr18_-_58369579
|
14.409
|
NM_010181
|
Fbn2
|
fibrillin 2
|
|
chr11_+_32355355
|
14.358
|
NM_173784
|
Ubtd2
|
ubiquitin domain containing 2
|
|
chr10_+_21698470
|
14.316
|
NM_001161847
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_+_113384418
|
14.155
|
NM_001097621
|
Kif26a
|
kinesin family member 26A
|
|
chr10_+_87323855
|
13.362
|
NM_001111274
NM_001111276
NM_184052
|
Igf1
|
insulin-like growth factor 1
|
|
chr9_+_108855789
|
13.236
|
NM_007738
|
Col7a1
|
collagen, type VII, alpha 1
|
|
chr6_-_128093543
|
13.142
|
NM_175414
|
Tspan9
|
tetraspanin 9
|
|
chr2_+_35413977
|
13.108
|
NM_001114125
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr3_+_67696141
|
13.016
|
NM_001113419
NM_177585
|
Iqcj-schip1
Iqcj
|
IQ motif containing J-schwannomin interacting protein 1 read-through transcript
IQ motif containing J
|
|
chr18_-_52689279
|
12.988
|
NM_010728
|
Lox
|
lysyl oxidase
|
|
chr7_-_16552844
|
12.940
|
NM_153068
|
Ehd2
|
EH-domain containing 2
|
|
chr4_-_82151180
|
12.813
|
NM_001113209
NM_001113210
NM_008687
|
Nfib
|
nuclear factor I/B
|
|
chr5_-_135223110
|
12.758
|
NM_007925
|
Eln
|
elastin
|
|
chr4_+_137024656
|
12.603
|
NM_008305
|
Hspg2
|
perlecan (heparan sulfate proteoglycan 2)
|
|
chr6_+_135312948
|
12.598
|
NM_010128
|
Emp1
|
epithelial membrane protein 1
|
|
chr7_-_141416778
|
12.359
|
NM_007400
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha)
|
|
chr9_-_83699706
|
12.228
|
NM_001145974
NM_148941
|
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr7_+_133968166
|
12.103
|
NM_001146347
|
Fam57b
|
family with sequence similarity 57, member B
|
|
chr18_+_53405310
|
12.003
|
NM_029394
|
Snx24
|
sorting nexing 24
|
|
chr4_+_47220883
|
11.645
|
NM_009928
|
Col15a1
|
collagen, type XV, alpha 1
|
|
chr11_-_78349594
|
11.517
|
NM_001159392
NM_009395
|
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr10_+_21601968
|
11.471
|
NM_001161845
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_14863507
|
11.261
|
NM_007606
|
Car3
|
carbonic anhydrase 3
|
|
chr1_+_24184448
|
10.980
|
NM_007740
|
Col9a1
|
collagen, type IX, alpha 1
|
|
chr10_+_21705909
|
10.847
|
NM_001161848
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr15_-_97835044
|
10.836
|
NM_001113515
NM_031163
|
Col2a1
|
collagen, type II, alpha 1
|
|
chr4_+_97444312
|
10.834
|
NM_001122953
NM_010905
|
Nfia
|
nuclear factor I/A
|
|
chr19_-_31839462
|
10.627
|
NM_001013833
|
Prkg1
|
protein kinase, cGMP-dependent, type I
|
|
chr6_+_17231334
|
10.537
|
NM_016900
|
Cav2
|
caveolin 2
|
|
chr3_-_86978431
|
10.444
|
NM_001170985
NM_130867
|
Kirrel
|
kin of IRRE like (Drosophila)
|
|
chr13_+_81022644
|
10.378
|
NM_001042591
|
Arrdc3
|
arrestin domain containing 3
|
|
chr10_-_62342761
|
10.371
|
NM_027384
|
Tet1
|
tet oncogene 1
|
|
chr19_+_47089168
|
10.082
|
NM_146100
|
Ina
|
internexin neuronal intermediate filament protein, alpha
|
|
chr8_+_71404452
|
9.845
|
NM_008509
|
Lpl
|
lipoprotein lipase
|
|
chr10_+_21713669
|
9.836
|
NM_001161850
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr4_-_82666973
|
9.801
|
NM_177863
|
Frem1
|
Fras1 related extracellular matrix protein 1
|
|
chr7_+_133966816
|
9.764
|
NM_029978
|
Fam57b
|
family with sequence similarity 57, member B
|
|
chr13_+_9275741
|
9.663
|
NM_001081426
|
Dip2c
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr16_+_21204820
|
9.603
|
NM_010143
|
Ephb3
|
Eph receptor B3
|
|
chrX_+_133357268
|
9.415
|
NM_011123
|
Plp1
|
proteolipid protein (myelin) 1
|
|
chr19_-_47538898
|
8.982
|
NM_001164717
NM_008018
|
Sh3pxd2a
|
SH3 and PX domains 2A
|
|
chr1_-_155179844
|
8.930
|
NM_010683
|
Lamc1
|
laminin, gamma 1
|
|
chr19_-_10944196
|
8.875
|
NM_133804
|
Tmem132a
|
transmembrane protein 132A
|
|
chr1_-_44175585
|
8.867
|
NM_023645
|
Kdelc1
|
KDEL (Lys-Asp-Glu-Leu) containing 1
|
|
chr15_+_66722913
|
8.859
|
NM_018865
|
Wisp1
|
WNT1 inducible signaling pathway protein 1
|
|
chr4_+_136025675
|
8.822
|
NM_024452
|
Luzp1
|
leucine zipper protein 1
|
|
chr5_+_96802973
|
8.693
|
NM_175473
|
Fras1
|
Fraser syndrome 1 homolog (human)
|
|
chr6_+_100654716
|
8.679
|
NM_198612
|
Gxylt2
|
glucoside xylosyltransferase 2
|
|
chr4_-_41587334
|
8.659
|
NM_027147
|
Enho
|
energy homeostasis associated
|
|
chr1_-_145624156
|
8.620
|
NM_013835
|
Trove2
|
TROVE domain family, member 2
|
|
chr17_-_63230665
|
8.566
|
NM_010109
NM_207654
|
Efna5
|
ephrin A5
|
|
chr11_-_86807216
|
8.560
|
NM_001005341
|
Ypel2
|
yippee-like 2 (Drosophila)
|
|
chr18_-_36675458
|
8.538
|
NM_010415
|
Hbegf
|
heparin-binding EGF-like growth factor
|
|
chr8_+_85387234
|
8.446
|
NM_177378
|
Rnf150
|
ring finger protein 150
|
|
chr15_-_78360039
|
8.414
|
NM_028331
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6
|
|
chr2_+_172376490
|
8.396
|
NM_001159696
|
Tcfap2c
|
transcription factor AP-2, gamma
|
|
chr8_+_98226914
|
8.380
|
NM_001195006
NM_145602
|
Ndrg4
|
N-myc downstream regulated gene 4
|
|
chr11_-_90252228
|
8.331
|
NM_172563
|
Hlf
|
hepatic leukemia factor
|
|
chr3_+_67178016
|
8.313
|
NM_001039543
NM_010801
|
Mlf1
|
myeloid leukemia factor 1
|
|
chr16_-_26371711
|
8.312
|
NM_016674
|
Cldn1
|
claudin 1
|
|
chr5_+_73872293
|
8.260
|
NM_001190734
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr2_-_102026486
|
8.169
|
NM_178886
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3
|
|
chr14_+_47502637
|
8.169
|
NM_001037221
NM_028966
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr8_+_87583954
|
8.077
|
NM_026791
|
Fbxw9
|
F-box and WD-40 domain protein 9
|
|
chr13_+_93381086
|
7.765
|
NM_172588
|
Serinc5
|
serine incorporator 5
|
|
chr4_-_82698070
|
7.750
|
NM_001198811
|
Frem1
|
Fras1 related extracellular matrix protein 1
|
|
chr7_+_133960398
|
7.750
|
NM_026884
|
Fam57b
|
family with sequence similarity 57, member B
|
|
chr6_+_54553140
|
7.742
|
NM_001001335
|
Plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8
|
|
chr13_-_43266716
|
7.556
|
NM_025935
|
Tbc1d7
|
TBC1 domain family, member 7
|
|
chr4_+_148919218
|
7.460
|
NM_001141930
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chrX_-_137908618
|
7.315
|
NM_053185
|
Col4a6
|
collagen, type IV, alpha 6
|
|
chr12_-_15823590
|
7.312
|
NM_144551
|
Trib2
|
tribbles homolog 2 (Drosophila)
|
|
chr4_+_148892231
|
7.297
|
NM_023465
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chr1_-_107253245
|
7.054
|
NM_001160368
NM_178779
|
Rnf152
|
ring finger protein 152
|
|
chr4_+_62876389
|
6.998
|
NM_025685
|
Col27a1
|
collagen, type XXVII, alpha 1
|
|
chr18_+_37089837
|
6.996
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chr7_+_54305973
|
6.961
|
NM_026436
|
Tmem86a
|
transmembrane protein 86A
|
|
chr18_+_37098858
|
6.862
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr18_+_37179802
|
6.855
|
NM_138663
|
Pcdha12
|
protocadherin alpha 12
|
|
chr12_-_77923303
|
6.807
|
NM_134050
|
Rab15
|
RAB15, member RAS oncogene family
|
|
chr9_-_20619426
|
6.757
|
NM_016919
|
Col5a3
|
collagen, type V, alpha 3
|
|
chr2_+_172375092
|
6.681
|
NM_009335
|
Tcfap2c
|
transcription factor AP-2, gamma
|
|
chr18_+_23962468
|
6.680
|
NM_153058
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr13_-_25361773
|
6.654
|
NM_009513
|
Nrsn1
|
neurensin 1
|
|
chr14_+_21748646
|
6.545
|
NM_009502
|
Vcl
|
vinculin
|
|
chr6_+_54545104
|
6.426
|
NM_001164361
|
Plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8
|
|
chr2_+_67955769
|
6.405
|
NM_020283
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr15_-_100325669
|
6.352
|
NM_153407
|
Csrnp2
|
cysteine-serine-rich nuclear protein 2
|
|
chr11_+_45869473
|
6.330
|
NM_009616
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta)
|
|
chr5_+_73882264
|
6.310
|
NM_001190733
NM_178896
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr3_-_117063793
|
6.294
|
NM_177664
|
D3Bwg0562e
|
DNA segment, Chr 3, Brigham & Women's Genetics 0562 expressed
|
|
chr8_+_28134259
|
6.245
|
NM_153592
|
Erlin2
|
ER lipid raft associated 2
|
|
chr4_+_153544821
|
6.210
|
NM_001162977
|
Megf6
|
multiple EGF-like-domains 6
|
|
chr4_+_133871031
|
6.111
|
NM_001039048
|
Trim63
|
tripartite motif-containing 63
|
|
chr14_-_21613162
|
6.097
|
NM_001039138
NM_001039139
NM_178597
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chrX_-_141921588
|
6.040
|
NM_153319
|
Amot
|
angiomotin
|
|
chrX_-_92853721
|
6.027
|
NM_001003916
|
Zc4h2
|
zinc finger, C4H2 domain containing
|
|
chr7_+_99890204
|
6.009
|
NM_029494
|
Rab30
|
RAB30, member RAS oncogene family
|
|
chr17_-_68353397
|
5.858
|
NM_172964
|
Arhgap28
|
Rho GTPase activating protein 28
|
|
chr9_-_57315800
|
5.843
|
NM_020270
|
Scamp5
|
secretory carrier membrane protein 5
|
|
chrX_-_149204003
|
5.826
|
NM_172441
|
Shroom2
|
shroom family member 2
|
|
chr12_-_71448600
|
5.770
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr18_+_34380575
|
5.765
|
NM_007462
|
Apc
|
adenomatosis polyposis coli
|
|
chr1_+_63492447
|
5.738
|
NM_001177600
NM_011780
|
Adam23
|
a disintegrin and metallopeptidase domain 23
|
|
chr4_+_13670425
|
5.718
|
NM_001111026
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_+_62021728
|
5.712
|
NM_175090
|
Slc31a1
|
solute carrier family 31, member 1
|
|
chr1_+_17591981
|
5.687
|
NM_053191
|
Pi15
|
peptidase inhibitor 15
|
|
chr16_-_38800293
|
5.645
|
NM_178924
|
Upk1b
|
uroplakin 1B
|
|
chr14_-_46149660
|
5.580
|
NM_146054
|
Fermt2
|
fermitin family homolog 2 (Drosophila)
|
|
chr18_+_37133455
|
5.573
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chr2_+_155207581
|
5.558
|
NM_178111
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2
|
|
chr1_-_152840432
|
5.534
|
NM_001024720
|
Hmcn1
|
hemicentin 1
|
|
chr10_-_13194201
|
5.527
|
NM_001195065
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr11_-_72225084
|
5.515
|
NM_177776
|
Smtnl2
|
smoothelin-like 2
|
|
chrX_+_5624624
|
5.470
|
NM_021431
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chrX_-_133276147
|
5.442
|
NM_019768
|
Morf4l2
|
mortality factor 4 like 2
|
|
chrX_+_20265590
|
5.440
|
NM_011049
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr12_+_112812710
|
5.435
|
NM_021516
NM_022801
|
Mark3
|
MAP/microtubule affinity-regulating kinase 3
|
|
chr9_+_121669121
|
5.410
|
NM_001166644
|
Zfp651
|
zinc finger protein 651
|
|
chr1_+_141318927
|
5.386
|
NM_172643
|
Zbtb41
|
zinc finger and BTB domain containing 41 homolog
|
|
chr4_+_13711928
|
5.236
|
NM_009822
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_17717497
|
5.235
|
NM_031402
|
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr18_+_37105758
|
5.157
|
NM_138662
|
Pcdha3
|
protocadherin alpha 3
|
|
chr10_+_79256298
|
5.133
|
NM_001161747
NM_023128
|
Palm
|
paralemmin
|
|
chrX_-_101017286
|
5.125
|
NM_009197
|
Slc16a2
|
solute carrier family 16 (monocarboxylic acid transporters), member 2
|
|
chr18_+_37120093
|
5.102
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr17_-_46162111
|
5.095
|
NM_001110266
NM_001110267
NM_001110268
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr15_+_79346819
|
5.079
|
NM_134090
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr6_-_89312545
|
5.073
|
NM_008881
|
Plxna1
|
plexin A1
|
|
chr9_+_56712850
|
5.047
|
NM_139001
|
Cspg4
|
chondroitin sulfate proteoglycan 4
|
|
chr4_-_57156022
|
5.039
|
NM_019427
|
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b
|
|
chr13_-_102538106
|
5.018
|
NM_001077495
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)
|
|
chr9_-_58388489
|
4.942
|
NM_133983
|
Cd276
|
CD276 antigen
|
|
chr18_+_37157533
|
4.908
|
NM_138661
|
Pcdha9
|
protocadherin alpha 9
|