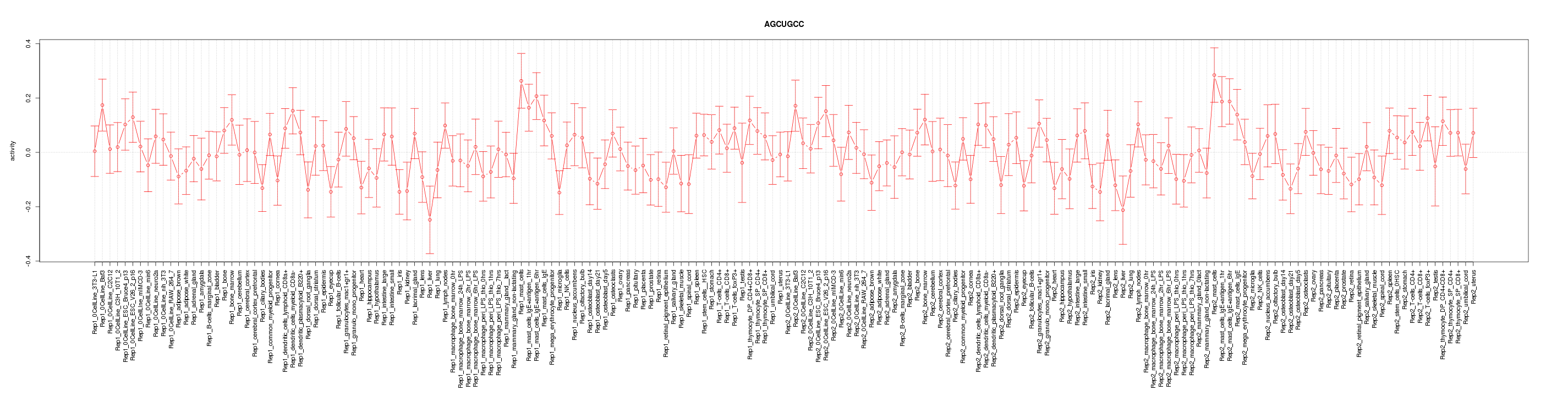
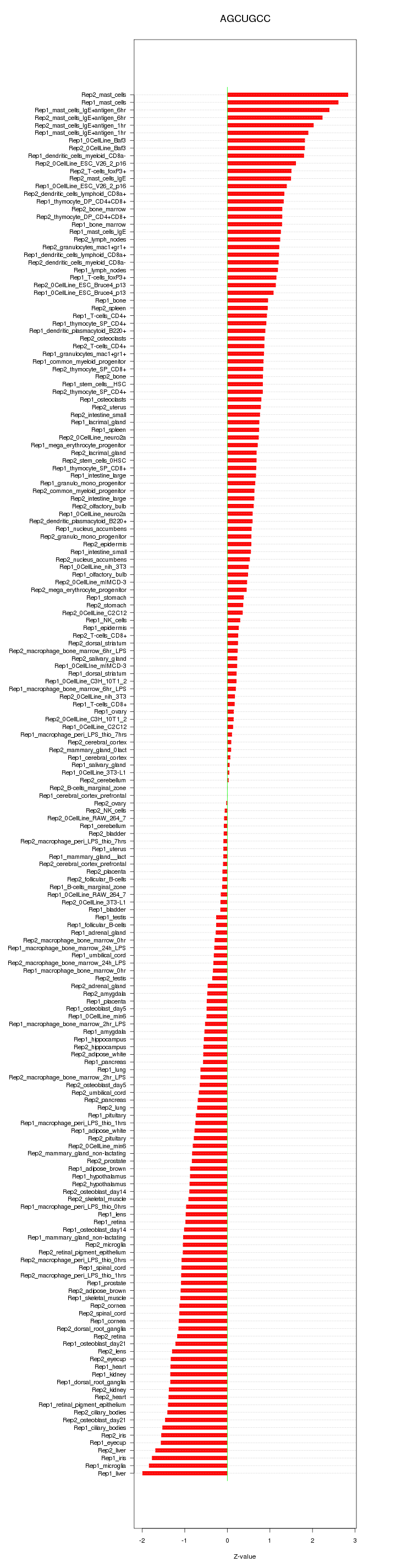
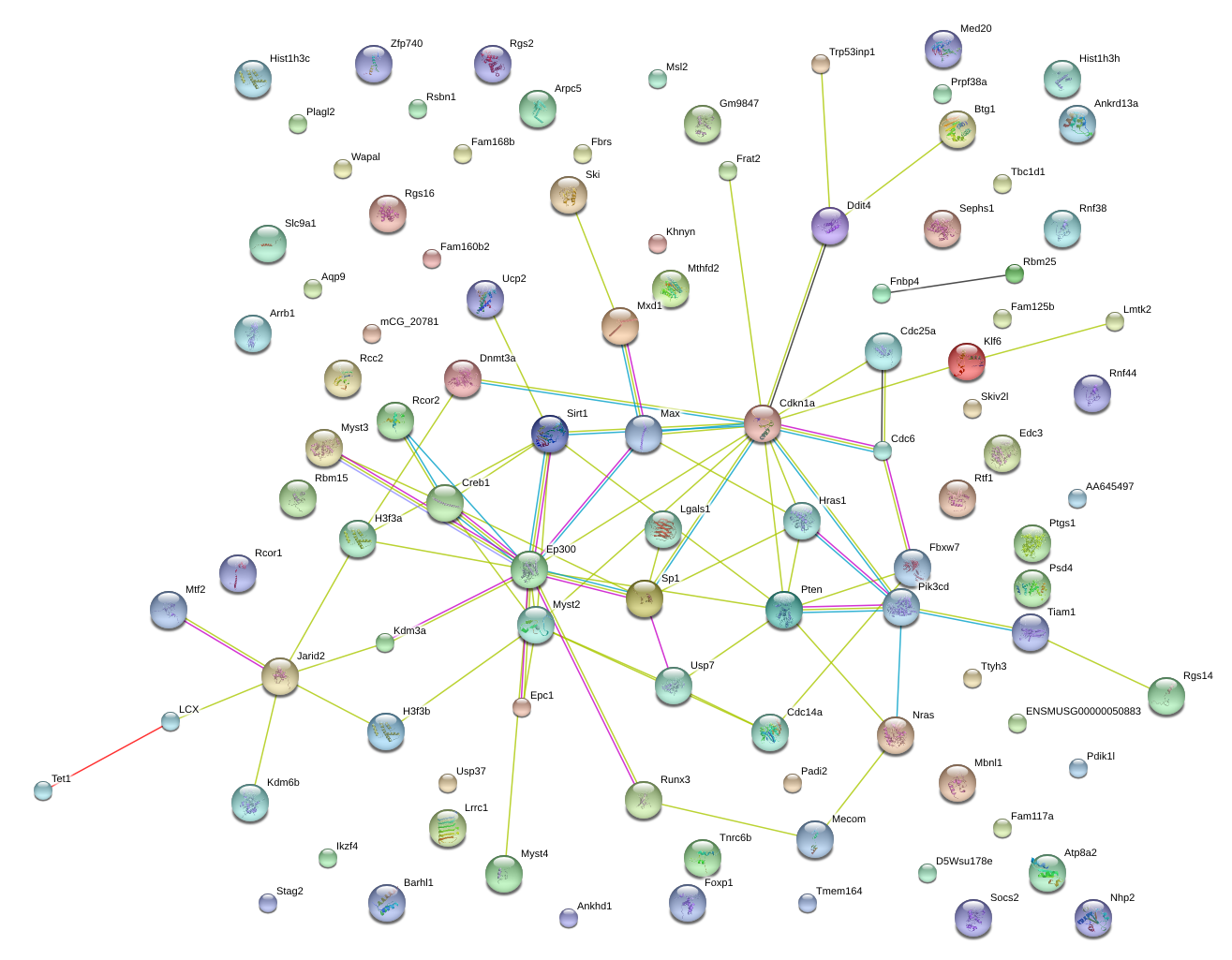
|
chr11_+_98769464
|
14.197
|
NM_011799
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr11_+_98769162
|
13.950
|
NM_001025779
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr4_+_140257340
|
11.926
|
NM_173867
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr10_+_96079634
|
11.284
|
NM_007569
|
Btg1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr6_-_83267540
|
10.558
|
NM_008638
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase
|
|
chr16_-_89818596
|
9.384
|
NM_001145887
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr19_-_41922577
|
9.096
|
NM_177603
|
Frat2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr4_+_11083585
|
8.146
|
NM_021897
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1
|
|
chr9_+_100978450
|
7.899
|
NM_001100451
|
Msl2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr1_-_74590801
|
7.641
|
NM_176972
|
Usp37
|
ubiquitin specific peptidase 37
|
|
chr9_+_57556346
|
7.604
|
NM_153799
|
Edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae)
|
|
chr5_+_115224911
|
7.431
|
NM_026718
|
Ankrd13a
|
ankyrin repeat domain 13a
|
|
chrX_+_139115908
|
7.369
|
NM_177592
|
Tmem164
|
transmembrane protein 164
|
|
chr2_+_36085945
|
6.896
|
NM_008969
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1
|
|
chr13_+_5860701
|
6.855
|
NM_011803
|
Klf6
|
Kruppel-like factor 6
|
|
chr19_+_32832002
|
6.809
|
NM_008960
|
Pten
|
phosphatase and tensin homolog
|
|
chr12_+_112278008
|
6.500
|
NM_198023
|
Rcor1
|
REST corepressor 1
|
|
chr10_-_59414516
|
6.043
|
NM_029083
|
Ddit4
|
DNA-damage-inducible transcript 4
|
|
chr6_-_86619001
|
5.834
|
NM_010751
|
Mxd1
|
MAX dimerization protein 1
|
|
chr11_-_115885714
|
5.723
|
NM_008211
|
H3f3b
|
H3 histone, family 3B
|
|
chr4_-_133843139
|
5.683
|
NM_001163794
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chr11_+_51433236
|
5.542
|
NM_026631
|
Nhp2
|
NHP2 ribonucleoprotein homolog (yeast)
|
|
chr16_-_89960987
|
5.497
|
NM_009384
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr9_-_77391865
|
5.482
|
NM_172528
|
Lrrc1
|
leucine rich repeat containing 1
|
|
chr15_+_80541675
|
5.453
|
NM_144812
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chr4_-_133843746
|
5.322
|
NM_146156
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chr13_+_44826121
|
5.285
|
NM_021878
|
Jarid2
|
jumonji, AT rich interactive domain 2
|
|
chr1_-_145851257
|
5.175
|
NM_009061
|
Rgs2
|
regulator of G-protein signaling 2
|
|
chr5_+_108494692
|
5.110
|
NM_013827
|
Mtf2
|
metal response element binding transcription factor 2
|
|
chr6_-_98978185
|
5.092
|
NM_001197322
|
Foxp1
|
forkhead box P1
|
|
chr15_+_102236709
|
5.076
|
NM_013672
|
Sp1
|
trans-acting transcription factor 1
|
|
chr10_-_128082881
|
5.041
|
NM_011772
|
Ikzf4
|
IKAROS family zinc finger 4
|
|
chr5_+_64551449
|
4.968
|
NM_019636
|
Tbc1d1
|
TBC1 domain family, member 1
|
|
chr11_-_69227176
|
4.949
|
NM_001017426
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B
|
|
chr13_+_55471092
|
4.934
|
NM_016758
|
Rgs14
|
regulator of G-protein signaling 14
|
|
chr2_+_90585526
|
4.848
|
NM_018828
|
Fnbp4
|
formin binding protein 4
|
|
chr15_+_78757148
|
4.827
|
NM_008495
|
Lgals1
|
lectin, galactose binding, soluble 1
|
|
chr4_-_149050154
|
4.818
|
NM_001164051
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr16_-_89974943
|
4.795
|
NM_001145886
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr6_-_99216494
|
4.731
|
NM_001197321
|
Foxp1
|
forkhead box P1
|
|
chr6_-_71582871
|
4.707
|
NM_001038695
|
Kdm3a
|
lysine (K)-specific demethylase 3A
|
|
chr16_-_8738377
|
4.696
|
NM_001003918
|
Usp7
|
ubiquitin specific peptidase 7
|
|
chr4_+_140462254
|
4.666
|
NM_008812
|
Padi2
|
peptidyl arginine deiminase, type II
|
|
chr1_+_64579358
|
4.525
|
NM_001037726
NM_009952
NM_133828
|
Creb1
|
cAMP responsive element binding protein 1
|
|
chr15_+_80629020
|
4.522
|
NM_177124
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chr4_-_149072755
|
4.440
|
NM_001164049
NM_001164050
NM_008840
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chrX_+_39503858
|
4.389
|
NM_021465
|
Stag2
|
stromal antigen 2
|
|
chr4_-_149037392
|
4.277
|
NM_001164052
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr2_-_153067043
|
4.118
|
NM_018807
|
Plagl2
|
pleiomorphic adenoma gene-like 2
|
|
chr1_+_154613622
|
4.098
|
NM_026369
|
Arpc5
|
actin related protein 2/3 complex, subunit 5
|
|
chr6_-_99385338
|
4.080
|
NM_053202
|
Foxp1
|
forkhead box P1
|
|
chr5_-_141124884
|
4.062
|
NM_175274
|
Ttyh3
|
tweety homolog 3 (Drosophila)
|
|
chr18_-_6490848
|
4.060
|
NM_027497
|
Epc1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr7_+_106683983
|
3.988
|
NM_177231
NM_178220
|
Arrb1
|
arrestin, beta 1
|
|
chr6_-_71582562
|
3.979
|
NM_173001
|
Kdm3a
|
lysine (K)-specific demethylase 3A
|
|
chr9_-_77392505
|
3.967
|
NM_001146048
|
Lrrc1
|
leucine rich repeat containing 1
|
|
chr3_+_60305108
|
3.893
|
NM_020007
|
Mbnl1
|
muscleblind-like 1 (Drosophila)
|
|
chr7_+_134628622
|
3.833
|
NM_010183
|
Fbrs
|
fibrosin
|
|
chr12_+_84973146
|
3.830
|
NM_027349
|
AA645497
Rbm25
|
expressed sequence AA645497
RNA binding motif protein 25
|
|
chr10_-_62801726
|
3.741
|
NM_001159590
NM_019812
NM_001159589
|
Sirt1
|
sirtuin 1 (silent mating type information regulation 2, homolog) 1 (S. cerevisiae)
|
|
chr11_+_95198329
|
3.713
|
NM_172543
|
Fam117a
|
family with sequence similarity 117, memberA
|
|
chr15_+_102038697
|
3.683
|
NM_153194
|
Zfp740
|
zinc finger protein 740
|
|
chr17_+_29230715
|
3.587
|
NM_007669
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21)
|
|
chr4_+_134676559
|
3.532
|
NM_019732
|
Runx3
|
runt related transcription factor 3
|
|
chr3_-_29912113
|
3.504
|
NM_007963
|
Mecom
|
MDS1 and EVI1 complex locus
|
|
chr8_+_23970006
|
3.497
|
NM_001081149
|
Myst3
|
MYST histone acetyltransferase (monocytic leukemia) 3
|
|
chr12_+_3806955
|
3.472
|
NM_007872
|
Dnmt3a
|
DNA methyltransferase 3A
|
|
chr9_+_109778051
|
3.430
|
NM_007658
|
Cdc25a
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr3_-_107135939
|
3.417
|
NM_001045807
|
Rbm15
|
RNA binding motif protein 15
|
|
chr2_-_33743465
|
3.375
|
NM_175184
|
Fam125b
|
family with sequence similarity 125, member B
|
|
chrX_+_39502588
|
3.374
|
NM_001077712
|
Stag2
|
stromal antigen 2
|
|
chr18_+_36823713
|
3.367
|
NM_201256
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr4_-_154596628
|
3.343
|
NM_011385
|
Ski
|
ski sarcoma viral oncogene homolog (avian)
|
|
chr4_-_149075697
|
3.321
|
NM_001029837
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr12_+_3891743
|
3.283
|
NM_153743
|
Dnmt3a
|
DNA methyltransferase 3A
|
|
chr14_+_56503783
|
3.259
|
NM_027143
|
Khnyn
|
KH and NYN domain containing
|
|
chr18_-_6516073
|
3.247
|
NM_007935
|
Epc1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr3_+_102862151
|
3.229
|
NM_010937
|
Nras
|
neuroblastoma ras oncogene
|
|
chr14_+_22319075
|
3.125
|
NM_017479
|
Myst4
|
MYST histone acetyltransferase monocytic leukemia 4
|
|
chr1_+_161974503
|
3.113
|
NM_001162896
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene
|
|
chr10_-_94878845
|
3.032
|
NM_001168655
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr13_-_54789148
|
3.020
|
NM_001146025
NM_001146026
NM_134064
|
Rnf44
|
ring finger protein 44
|
|
chr2_-_28771940
|
2.998
|
NM_019446
NM_001164186
|
Barhl1
|
BarH-like 1 (Drosophila)
|
|
chr3_+_84756090
|
2.991
|
NM_080428
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr2_+_24240916
|
2.890
|
NM_177611
|
Psd4
|
pleckstrin and Sec7 domain containing 4
|
|
chr2_+_112219335
|
2.863
|
NM_024254
|
2410042D21Rik
|
RIKEN cDNA 2410042D21 gene
|
|
chr14_+_35487052
|
2.854
|
NM_001004436
|
Wapal
|
wings apart-like homolog (Drosophila)
|
|
chr7_+_107641853
|
2.784
|
NM_011671
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier)
|
|
chr15_+_81416643
|
2.755
|
NM_177821
|
Ep300
|
E1A binding protein p300
|
|
chr9_-_71011053
|
2.738
|
NM_022026
|
Aqp9
|
aquaporin 9
|
|
chr10_-_62342761
|
2.707
|
NM_027384
|
Tet1
|
tet oncogene 1
|
|
chr3_-_116126932
|
2.676
|
NM_001080818
NM_001173553
|
Cdc14a
|
CDC14 cell division cycle 14 homolog A (S. cerevisiae)
|
|
chr4_-_44181148
|
2.672
|
NM_001038993
|
Rnf38
|
ring finger protein 38
|
|
chr5_+_30559138
|
2.649
|
NM_027652
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr4_+_132925684
|
2.596
|
NM_016981
|
Slc9a1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr17_+_47748530
|
2.588
|
NM_020048
|
Med20
|
mediator complex subunit 20
|
|
chr3_+_84619399
|
2.543
|
NM_001177773
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr12_-_78063011
|
2.542
|
NM_001146176
NM_008558
|
Max
|
Max protein
|
|
chr2_+_4802609
|
2.539
|
NM_175400
|
Sephs1
|
selenophosphate synthetase 1
|
|
chr5_+_144861227
|
2.522
|
NM_001081109
|
Lmtk2
|
lemur tyrosine kinase 2
|
|
chr1_-_34899878
|
2.522
|
NM_001160235
NM_001160236
NM_174997
|
Fam168b
|
family with sequence similarity 168, member B
|
|
chr11_-_95171521
|
2.484
|
NM_001195003
NM_001195004
NM_177619
|
Myst2
|
MYST histone acetyltransferase 2
|
|
chr14_-_70999625
|
2.446
|
NM_194345
|
Fam160b2
|
family with sequence similarity 160, member B2
|
|
chr2_+_119500777
|
2.442
|
NM_030112
|
Rtf1
|
Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr4_-_108251843
|
2.413
|
NM_172697
|
Prpf38a
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing A
|
|
chr3_+_103718042
|
2.359
|
NM_172684
|
Rsbn1
|
rosbin, round spermatid basic protein 1
|
|
chr1_-_194956399
|
2.354
|
NM_145415
|
Diexf
|
digestive organ expansion factor homolog (zebrafish)
|
|
chr14_+_56384798
|
2.345
|
NM_008519
|
Ltb4r1
|
leukotriene B4 receptor 1
|
|
chr1_-_179178874
|
2.332
|
NM_011785
|
Akt3
|
thymoma viral proto-oncogene 3
|
|
chr17_+_29227908
|
2.317
|
NM_001111099
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21)
|
|
chr8_+_93352723
|
2.308
|
NM_177224
|
Chd9
|
chromodomain helicase DNA binding protein 9
|
|
chr11_+_72861327
|
2.281
|
NM_025818
|
1200014J11Rik
|
RIKEN cDNA 1200014J11 gene
|
|
chr1_-_175297778
|
2.249
|
NM_053199
|
Cadm3
|
cell adhesion molecule 3
|
|
chr11_+_72503487
|
2.240
|
NM_009671
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr6_+_120616386
|
2.224
|
NM_001128151
|
Cecr2
|
cat eye syndrome chromosome region, candidate 2 homolog (human)
|
|
chr4_+_59202363
|
2.207
|
NM_011673
|
Ugcg
|
UDP-glucose ceramide glucosyltransferase
|
|
chr16_-_45654180
|
2.199
|
NM_145389
|
BC016579
|
cDNA sequence, BC016579
|
|
chr4_+_122673318
|
2.182
|
NM_008506
|
Mycl1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr19_-_60656627
|
2.135
|
NM_001172096
NM_001172097
NM_030197
|
2700078E11Rik
|
RIKEN cDNA 2700078E11 gene
|
|
chr4_+_128867038
|
2.081
|
NM_134151
|
Yars
|
tyrosyl-tRNA synthetase
|
|
chr10_-_94879435
|
2.079
|
NM_007706
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr6_-_37249957
|
2.070
|
NM_001081206
|
Dgki
|
diacylglycerol kinase, iota
|
|
chr3_+_84668066
|
2.063
|
NM_001177774
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr11_-_97605846
|
2.052
|
NM_054051
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr2_+_32426348
|
2.049
|
NM_010073
|
Dpm2
|
dolichol-phosphate (beta-D) mannosyltransferase 2
|
|
chr9_-_57682040
|
2.038
|
NM_019689
|
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr7_-_87550315
|
2.022
|
NM_001081454
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr10_-_94877824
|
1.979
|
NM_001168657
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr17_+_28828286
|
1.967
|
NM_001168508
NM_001168514
NM_011951
|
Mapk14
|
mitogen-activated protein kinase 14
|
|
chr6_-_32538191
|
1.948
|
NM_175750
|
Plxna4
|
plexin A4
|
|
chr2_-_160738055
|
1.936
|
NM_182840
|
Emilin3
|
elastin microfibril interfacer 3
|
|
chr3_+_98944990
|
1.906
|
NM_027462
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial)
|
|
chr18_-_39646734
|
1.900
|
NM_008173
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1
|
|
chr6_+_54766562
|
1.877
|
NM_199143
|
Znrf2
|
zinc and ring finger 2
|
|
chr12_-_4234293
|
1.868
|
NM_134046
|
Cenpo
|
centromere protein O
|
|
chr2_+_75497282
|
1.841
|
NM_053263
NM_146130
NM_198090
|
Hnrnpa3
Gm6793
|
heterogeneous nuclear ribonucleoprotein A3
heterogeneous nuclear ribonucleoprotein A3 pseudogene
|
|
chr11_-_117830554
|
1.821
|
NM_007707
|
Socs3
|
suppressor of cytokine signaling 3
|
|
chr4_-_44180323
|
1.816
|
NM_175201
|
Rnf38
|
ring finger protein 38
|
|
chr10_+_19654195
|
1.808
|
NM_008580
|
Map3k5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr5_-_124778459
|
1.788
|
NM_001081323
|
Mphosph9
|
M-phase phosphoprotein 9
|
|
chr15_-_96472392
|
1.788
|
NM_134086
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr3_-_133207209
|
1.776
|
NM_001040400
|
Tet2
|
tet oncogene family member 2
|
|
chr16_+_24448176
|
1.751
|
NM_001145952
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_-_128133261
|
1.737
|
NM_177411
|
Rab5b
|
RAB5B, member RAS oncogene family
|
|
chr2_-_65962685
|
1.716
|
NM_015736
|
Galnt3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3
|
|
chr13_+_55311142
|
1.668
|
NM_008739
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1
|
|
chr7_-_87547589
|
1.663
|
NM_011046
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr10_+_127814144
|
1.640
|
NM_172790
|
Ankrd52
|
ankyrin repeat domain 52
|
|
chr4_+_13678443
|
1.639
|
NM_001111027
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_+_156278399
|
1.616
|
NM_001135728
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr13_-_3917356
|
1.587
|
NM_019671
|
Net1
|
neuroepithelial cell transforming gene 1
|
|
chr3_+_88336316
|
1.583
|
NM_001029890
|
Mex3a
|
mex3 homolog A (C. elegans)
|
|
chr17_+_29797537
|
1.560
|
NM_028791
|
Ftsjd2
|
FtsJ methyltransferase domain containing 2
|
|
chr15_-_96473193
|
1.558
|
NM_001166458
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chrX_+_156065141
|
1.531
|
NM_001135727
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr5_-_68238654
|
1.507
|
NM_001038999
NM_009727
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chrX_+_156146532
|
1.493
|
NM_021389
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr10_-_94878117
|
1.486
|
NM_001168656
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr18_+_82644510
|
1.485
|
NM_001025245
NM_010777
|
Mbp
|
myelin basic protein
|
|
chr2_-_73052484
|
1.444
|
NM_025942
NM_030091
|
Ola1
|
Obg-like ATPase 1
|
|
chr19_+_5447546
|
1.441
|
NM_010235
|
Fosl1
|
fos-like antigen 1
|
|
chr13_-_54789548
|
1.422
|
NM_001146027
|
Rnf44
|
ring finger protein 44
|
|
chrX_-_71330931
|
1.417
|
NM_010788
NM_001081979
|
Mecp2
|
methyl CpG binding protein 2
|
|
chr7_+_109118356
|
1.380
|
NM_133947
|
Numa1
|
nuclear mitotic apparatus protein 1
|
|
chr1_+_88483674
|
1.351
|
NM_172974
|
Cops7b
|
COP9 (constitutive photomorphogenic) homolog, subunit 7b (Arabidopsis thaliana)
|
|
chr8_+_107064705
|
1.347
|
NM_053070
|
Car7
|
carbonic anhydrase 7
|
|
chr2_+_155601077
|
1.338
|
NM_010808
|
Mmp24
|
matrix metallopeptidase 24
|
|
chr11_+_105042843
|
1.332
|
NM_001112705
NM_011903
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis)
|
|
chr6_-_3713622
|
1.312
|
NM_001042725
|
Calcr
|
calcitonin receptor
|
|
chr4_-_139388882
|
1.307
|
NM_011039
|
Pax7
|
paired box gene 7
|
|
chr11_-_102742230
|
1.293
|
NM_001109995
NM_011431
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2
|
|
chr11_+_94491055
|
1.280
|
NM_133807
|
Lrrc59
|
leucine rich repeat containing 59
|
|
chr7_+_51565563
|
1.275
|
NM_001034115
|
Shank1
|
SH3/ankyrin domain gene 1
|
|
chr2_+_22751003
|
1.274
|
NM_019501
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1
|
|
chr17_-_84865455
|
1.257
|
NM_183021
|
Thada
|
thyroid adenoma associated
|
|
chr14_+_45970767
|
1.256
|
NM_019637
|
Styx
|
serine/threonine/tyrosine interaction protein
|
|
chr7_+_100023762
|
1.251
|
NM_028243
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C)
|
|
chr14_-_75347637
|
1.244
|
NM_001033439
|
Lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1
|
|
chr6_-_72389491
|
1.236
|
NM_145569
|
Mat2a
|
methionine adenosyltransferase II, alpha
|
|
chr7_-_54263783
|
1.223
|
NM_175318
|
Spty2d1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr16_+_17233664
|
1.222
|
NM_178922
|
Hic2
|
hypermethylated in cancer 2
|
|
chr4_-_35792385
|
1.214
|
NM_001166001
NM_175516
|
Lingo2
|
leucine rich repeat and Ig domain containing 2
|
|
chr4_-_151370212
|
1.212
|
NM_172705
|
Phf13
|
PHD finger protein 13
|
|
chr12_+_81891479
|
1.199
|
NM_178682
|
4933426M11Rik
|
RIKEN cDNA 4933426M11 gene
|
|
chr13_-_3892788
|
1.189
|
NM_001047159
|
Net1
|
neuroepithelial cell transforming gene 1
|
|
chr2_-_62249934
|
1.185
|
NM_001159543
NM_010074
|
Dpp4
|
dipeptidylpeptidase 4
|
|
chr19_-_5912757
|
1.183
|
NM_011262
|
Dpf2
|
D4, zinc and double PHD fingers family 2
|
|
chr18_-_67884185
|
1.179
|
NM_001127177
NM_008977
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2
|
|
chr1_+_192922288
|
1.176
|
NM_030060
|
Batf3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr15_+_83610434
|
1.171
|
NM_172610
|
Mpped1
|
metallophosphoesterase domain containing 1
|
|
chr1_-_57028171
|
1.142
|
NM_139146
|
Satb2
|
special AT-rich sequence binding protein 2
|
|
chr10_-_128258639
|
1.141
|
NM_024180
|
Ormdl2
|
ORM1-like 2 (S. cerevisiae)
|
|
chrX_+_147955201
|
1.139
|
NM_001113354
NM_177201
|
Phf8
|
PHD finger protein 8
|
|
chr9_+_106073255
|
1.131
|
NM_029896
|
Wdr82
|
WD repeat domain containing 82
|
|
chr11_+_68316520
|
1.110
|
NM_001004435
NM_001081566
|
Pik3r6
|
phosphoinositide-3-kinase, regulatory subunit 6
|
|
chr19_-_12870612
|
1.097
|
NM_053009
|
Zfp91
|
zinc finger protein 91
|
|
chr11_+_115794942
|
1.096
|
NM_020483
|
Sap30bp
|
SAP30 binding protein
|
|
chr17_+_26551909
|
1.091
|
NM_001081656
|
Neurl1b
|
neuralized homolog 1b (Drosophila)
|
|
chr2_-_156904966
|
1.057
|
NM_001164663
|
9830001H06Rik
|
RIKEN cDNA 9830001H06 gene
|
|
chr7_-_30290470
|
1.056
|
NM_001081028
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3
|