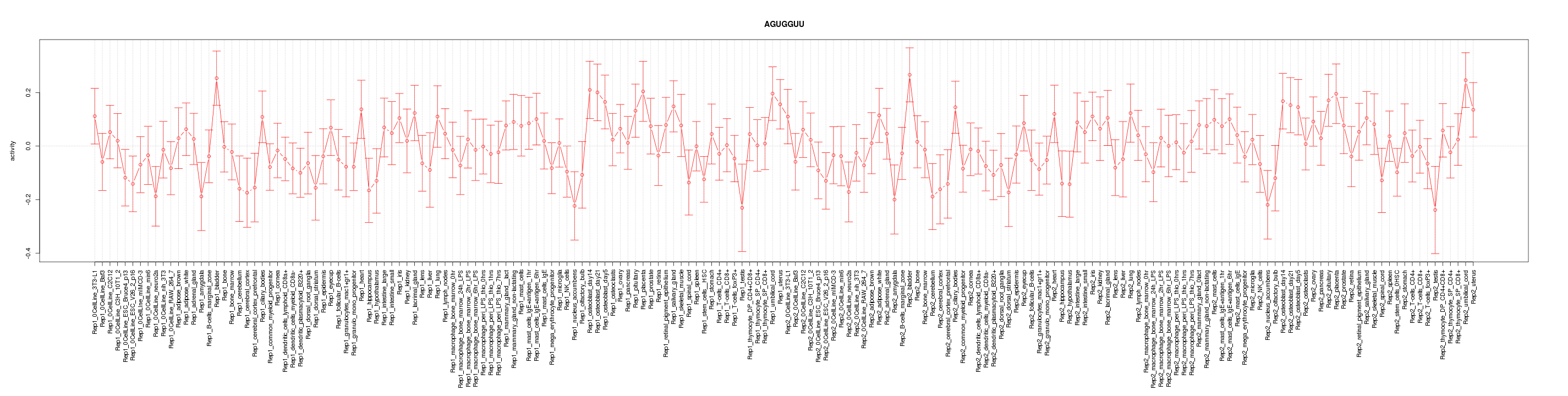
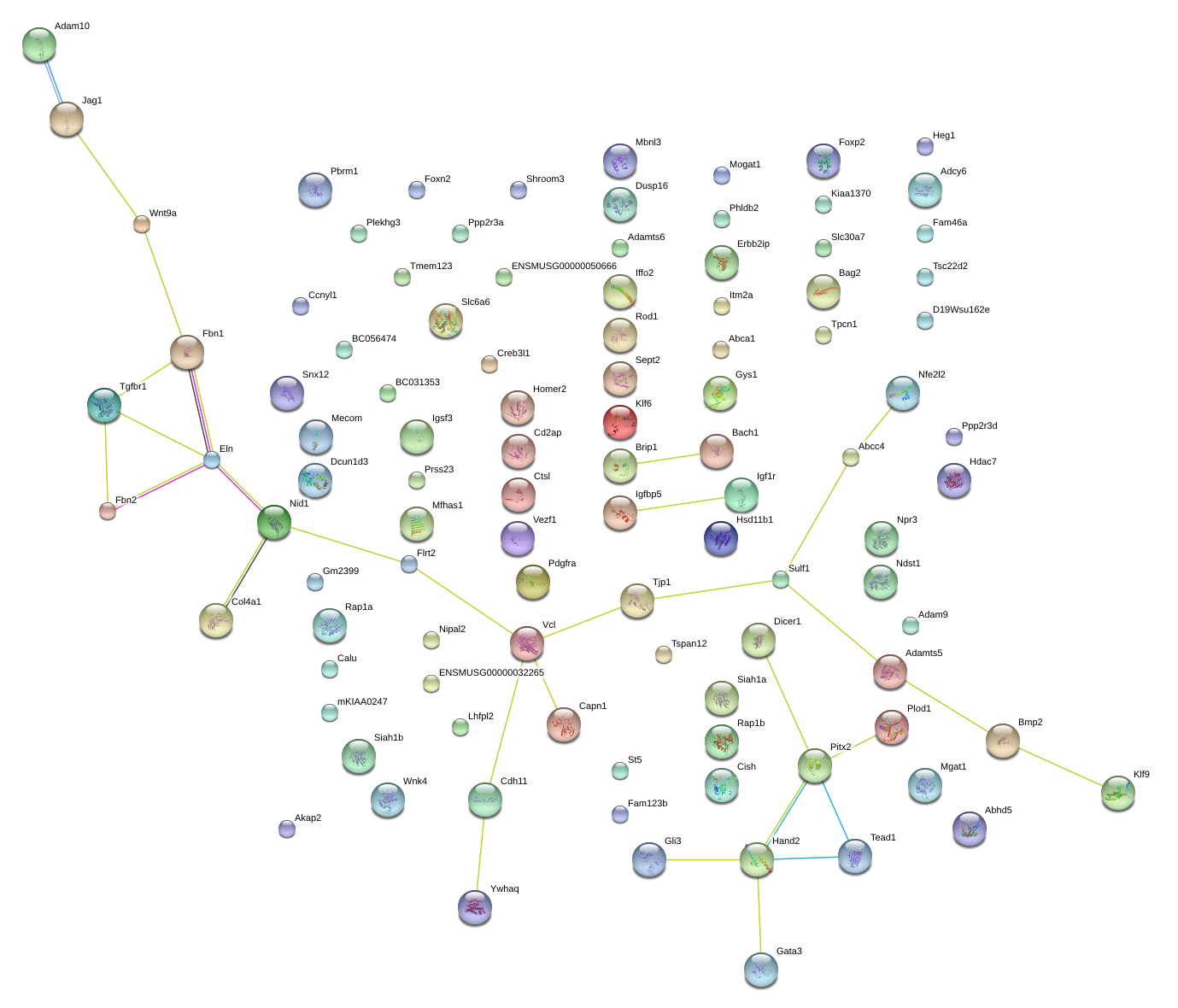
|
chr5_+_75552144
|
18.449
|
NM_001083316
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr2_-_91864276
|
17.818
|
NM_011957
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1
|
|
chr4_+_57867433
|
13.642
|
NM_001035533
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr5_+_75548309
|
12.988
|
NM_011058
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr4_+_57858096
|
12.893
|
NM_001035532
NM_009649
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr1_-_72921438
|
12.463
|
NM_010518
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr2_-_125332107
|
12.338
|
NM_007993
|
Fbn1
|
fibrillin 1
|
|
chr7_-_96666081
|
12.095
|
NM_029614
|
Prss23
|
protease, serine, 23
|
|
chr13_+_13529868
|
10.999
|
NM_010917
|
Nid1
|
nidogen 1
|
|
chrX_-_48559008
|
10.779
|
NM_134163
|
Mbnl3
|
muscleblind-like 3 (Drosophila)
|
|
chr8_-_105308910
|
10.747
|
NM_009866
|
Cdh11
|
cadherin 11
|
|
chr9_-_85220730
|
10.482
|
NM_001160378
NM_001160379
|
Fam46a
|
family with sequence similarity 46, member A
|
|
chr3_-_29912113
|
10.421
|
NM_007963
|
Mecom
|
MDS1 and EVI1 complex locus
|
|
chr15_-_11835393
|
10.274
|
NM_001039181
NM_008728
|
Npr3
|
natriuretic peptide receptor 3
|
|
chr16_-_85901362
|
9.958
|
NM_011782
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2)
|
|
chr7_-_116760660
|
8.949
|
NM_001001326
NM_029811
|
St5
|
suppression of tumorigenicity 5
|
|
chr7_+_119822831
|
8.428
|
NM_001166584
NM_009346
|
Tead1
|
TEA domain family member 1
|
|
chr14_+_21748646
|
7.980
|
NM_009502
|
Vcl
|
vinculin
|
|
chr4_-_147310804
|
7.618
|
NM_011122
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chr1_+_12682510
|
7.610
|
NM_001198565
NM_001198566
|
Sulf1
|
sulfatase 1
|
|
chr15_-_98438063
|
7.609
|
NM_007405
|
Adcy6
|
adenylate cyclase 6
|
|
chr2_+_133378934
|
7.295
|
NM_007553
|
Bmp2
|
bone morphogenetic protein 2
|
|
chr6_-_134742645
|
7.153
|
NM_001048054
NM_130447
|
Dusp16
|
dual specificity phosphatase 16
|
|
chr9_+_7764067
|
7.119
|
NM_133739
|
Tmem123
|
transmembrane protein 123
|
|
chr3_+_128916849
|
7.017
|
NM_001042502
|
Pitx2
|
paired-like homeodomain transcription factor 2
|
|
chr17_-_43013320
|
6.995
|
NM_009847
|
Cd2ap
|
CD2-associated protein
|
|
chr5_+_93327023
|
6.956
|
NM_001077596
|
Shroom3
|
shroom family member 3
|
|
chr1_+_12708582
|
6.923
|
NM_172294
|
Sulf1
|
sulfatase 1
|
|
chr9_-_101154160
|
6.733
|
NM_001161362
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr3_+_128902822
|
6.554
|
NM_011098
NM_001042504
|
Pitx2
|
paired-like homeodomain transcription factor 2
|
|
chr5_+_93238413
|
6.522
|
NM_001077595
|
Shroom3
|
shroom family member 3
|
|
chr15_-_34608458
|
6.236
|
NM_145469
|
Nipal2
|
NIPA-like domain containing 2
|
|
chr3_-_115710131
|
6.220
|
NM_023214
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr8_-_26127376
|
5.751
|
NM_007404
|
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma)
|
|
chr6_+_14851348
|
5.659
|
NM_053242
|
Foxp2
|
forkhead box P2
|
|
chr13_+_5860701
|
5.542
|
NM_011803
|
Klf6
|
Kruppel-like factor 6
|
|
chr6_+_15135505
|
5.517
|
NM_212435
|
Foxp2
|
forkhead box P2
|
|
chr7_+_119902897
|
5.473
|
NM_001166585
|
Tead1
|
TEA domain family member 1
|
|
chr15_-_97662101
|
5.452
|
NM_019572
|
Hdac7
|
histone deacetylase 7
|
|
chr4_+_47366162
|
5.395
|
NM_009370
|
Tgfbr1
|
transforming growth factor, beta receptor I
|
|
chr19_+_46673553
|
5.351
|
NM_001177812
NM_146099
|
D19Wsu162e
|
DNA segment, Chr 19, Wayne State University 162, expressed
|
|
chr13_-_64471609
|
5.350
|
NM_009984
|
Ctsl
|
cathepsin L
|
|
chr2_-_136942066
|
5.321
|
NM_013822
|
Jag1
|
jagged 1
|
|
chr19_+_23215704
|
5.224
|
NM_010638
|
Klf9
|
Kruppel-like factor 9
|
|
chr18_-_58369579
|
5.148
|
NM_010181
|
Fbn2
|
fibrillin 2
|
|
chr12_+_81891479
|
5.027
|
NM_178682
|
4933426M11Rik
|
RIKEN cDNA 4933426M11 gene
|
|
chr19_+_46697882
|
4.784
|
NM_001177813
|
D19Wsu162e
|
DNA segment, Chr 19, Wayne State University 162, expressed
|
|
chr7_-_72516109
|
4.715
|
NM_001163574
NM_009386
|
Tjp1
|
tight junction protein 1
|
|
chr4_-_59562054
|
4.712
|
NM_144904
NM_178164
|
Rod1
|
ROD1 regulator of differentiation 1 (S. pombe)
|
|
chr1_-_33814590
|
4.567
|
NM_145392
|
Bag2
|
BCL2-associated athanogene 2
|
|
chr9_-_101101334
|
4.554
|
NM_172144
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr10_-_117283020
|
4.497
|
NM_024457
|
Rap1b
|
RAS related protein 1b
|
|
chr6_+_91634056
|
4.355
|
NM_009320
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr13_+_105077914
|
4.336
|
NM_001081020
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6
|
|
chr5_-_121038602
|
3.849
|
NM_145853
|
Tpcn1
|
two pore channel 1
|
|
chr11_+_87881747
|
3.788
|
NM_016686
|
Vezf1
|
vascular endothelial zinc finger 1
|
|
chr1_+_95375503
|
3.753
|
NM_001159717
NM_001159718
NM_010891
NM_001159719
|
Sept2
|
septin 2
|
|
chr6_-_21802514
|
3.719
|
NM_173007
|
Tspan12
|
tetraspanin 12
|
|
chr7_+_52690164
|
3.719
|
NM_030678
|
Gys1
|
glycogen synthase 1, muscle
|
|
chr12_+_77634546
|
3.627
|
NM_153804
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr2_-_9800211
|
3.545
|
NM_008091
|
Gata3
|
GATA binding protein 3
|
|
chr16_+_33684551
|
3.504
|
NM_175256
|
Heg1
|
HEG homolog 1 (zebrafish)
|
|
chr9_+_70526702
|
3.504
|
NM_007399
|
Adam10
|
a disintegrin and metallopeptidase domain 10
|
|
chr7_+_75097142
|
3.465
|
NM_010513
|
Igf1r
|
insulin-like growth factor I receptor
|
|
chr9_+_122260733
|
3.310
|
NM_026179
|
Abhd5
|
abhydrolase domain containing 5
|
|
chrX_-_98417753
|
3.242
|
NM_001110310
NM_001110311
NM_018875
|
Snx12
|
sorting nexin 12
|
|
chr7_-_88851798
|
3.238
|
NM_001164086
NM_011983
|
Homer2
|
homer homolog 2 (Drosophila)
|
|
chr12_+_96930429
|
3.156
|
NM_201518
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr5_-_135223110
|
3.134
|
NM_007925
|
Eln
|
elastin
|
|
chr13_+_94827750
|
3.115
|
NM_172589
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2
|
|
chr13_+_15555483
|
3.110
|
NM_008130
|
Gli3
|
GLI-Kruppel family member GLI3
|
|
chr8_-_11312729
|
3.073
|
NM_009931
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr3_+_58219610
|
2.986
|
NM_001081229
|
Tsc22d2
|
TSC22 domain family, member 2
|
|
chr4_-_53172748
|
2.963
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr17_+_88840048
|
2.951
|
NM_180974
|
Foxn2
|
forkhead box N2
|
|
chr16_+_87699143
|
2.928
|
NM_007520
|
Bach1
|
BTB and CNC homology 1
|
|
chr3_+_101180976
|
2.889
|
NM_207205
|
Igsf3
|
immunoglobulin superfamily, member 3
|
|
chr16_-_45844409
|
2.874
|
NM_153412
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr13_-_104710374
|
2.851
|
NM_001005868
NM_021563
|
Erbb2ip
|
Erbb2 interacting protein
|
|
chr19_-_6015778
|
2.833
|
NM_001110504
|
Capn1
|
calpain 1
|
|
chrX_-_92640149
|
2.792
|
NM_175179
|
Fam123b
|
family with sequence similarity 123, member B
|
|
chr6_+_29298117
|
2.784
|
NM_007594
NM_184053
|
Calu
|
calumenin
|
|
chr14_+_33669941
|
2.757
|
NM_178791
|
E130203B14Rik
|
RIKEN cDNA E130203B14 gene
|
|
chr11_+_49057692
|
2.701
|
NM_001110148
NM_010794
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1
|
|
chr9_+_74800828
|
2.545
|
NM_001113283
|
BC031353
|
cDNA sequence BC031353
|
|
chr18_-_60872888
|
2.491
|
NM_008306
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr4_+_139131050
|
2.384
|
NM_183148
|
Iffo2
|
intermediate filament family orphan 2
|
|
chr14_+_31832323
|
2.374
|
NM_001081251
|
Pbrm1
|
polybromo 1
|
|
chr8_+_59799779
|
2.335
|
NM_010402
|
Hand2
|
heart and neural crest derivatives expressed transcript 2
|
|
chr8_+_36650839
|
2.325
|
NM_001081279
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr11_+_59120429
|
2.253
|
NM_139298
|
Wnt9a
|
wingless-type MMTV integration site 9A
|
|
chr7_-_127039189
|
2.244
|
NM_001163703
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr1_+_64737918
|
2.215
|
NM_001097644
|
Ccnyl1
|
cyclin Y-like 1
|
|
chr14_-_119105434
|
2.209
|
NM_001033336
NM_001163676
|
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4
|
|
chr2_-_75542552
|
2.191
|
NM_010902
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2
|
|
chr1_-_195090178
|
2.157
|
NM_008288
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1
|
|
chrX_-_104598651
|
2.120
|
NM_008409
|
Itm2a
|
integral membrane protein 2A
|
|
chr12_-_21423296
|
2.119
|
NM_011739
|
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr19_-_6015125
|
2.116
|
NM_007600
|
Capn1
|
calpain 1
|
|
chr11_+_101121880
|
2.065
|
NM_175638
|
Wnk4
|
WNK lysine deficient protein kinase 4
|
|
chr1_-_195068455
|
2.035
|
NM_001044751
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1
|
|
chr8_-_89269853
|
2.034
|
NM_009172
|
Siah1a
|
seven in absentia 1A
|
|
chr6_-_28211600
|
2.018
|
NM_001081678
|
Zfp800
|
zinc finger protein 800
|
|
chr10_-_13108775
|
2.008
|
NM_001195066
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr16_-_75766992
|
1.991
|
NM_030201
|
Hspa13
|
heat shock protein 70 family, member 13
|
|
chr15_-_77672472
|
1.954
|
NM_022410
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle
|
|
chr11_+_49064013
|
1.936
|
NM_001110150
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1
|
|
chr14_-_119076152
|
1.899
|
NM_001163675
|
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4
|
|
chr7_-_118226515
|
1.879
|
NM_001040131
NM_013507
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2
|
|
chr7_+_105987354
|
1.870
|
NM_009519
|
Wnt11
|
wingless-related MMTV integration site 11
|
|
chr9_+_74823917
|
1.853
|
NM_153584
|
BC031353
|
cDNA sequence BC031353
|
|
chr2_-_120389559
|
1.833
|
NM_011743
|
Zfp106
|
zinc finger protein 106
|
|
chr4_-_141161882
|
1.797
|
NM_133754
|
Fblim1
|
filamin binding LIM protein 1
|
|
chr11_+_49060978
|
1.785
|
NM_001110149
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1
|
|
chr14_+_49065971
|
1.764
|
NM_172600
|
6720456H20Rik
|
RIKEN cDNA 6720456H20 gene
|
|
chr18_+_56867378
|
1.755
|
NM_010721
|
Lmnb1
|
lamin B1
|
|
chr10_-_13043959
|
1.709
|
NM_001033257
NM_001195096
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr4_-_141155782
|
1.688
|
NM_001163256
|
Fblim1
|
filamin binding LIM protein 1
|
|
chr1_-_135557934
|
1.632
|
NM_144530
|
Zbed6
Zc3h11a
|
zinc finger, BED domain containing 6
zinc finger CCCH type containing 11A
|
|
chr14_+_55502261
|
1.516
|
NM_007537
|
Bcl2l2
|
BCL2-like 2
|
|
chr7_-_127038916
|
1.507
|
NM_173408
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr12_-_85749901
|
1.480
|
NM_001026214
NM_007647
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5
|
|
chr9_-_86465373
|
1.444
|
NM_001163746
NM_028352
|
Pgm3
|
phosphoglucomutase 3
|
|
chr2_+_164286416
|
1.438
|
NM_025575
|
Sys1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr1_-_174012771
|
1.435
|
NM_021607
|
Ncstn
|
nicastrin
|
|
chr10_-_13194201
|
1.427
|
NM_001195065
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr19_+_5038825
|
1.397
|
NM_175383
|
B3gnt1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr15_+_25552273
|
1.369
|
NM_019472
|
Myo10
|
myosin X
|
|
chr15_+_73578455
|
1.336
|
NM_008975
|
Ptp4a3
|
protein tyrosine phosphatase 4a3
|
|
chrX_+_137212566
|
1.313
|
NM_011845
|
Mid2
|
midline 2
|
|
chr7_+_87171113
|
1.312
|
NM_175433
|
Zfp710
|
zinc finger protein 710
|
|
chr3_+_9250566
|
1.287
|
NM_177660
|
Zbtb10
|
zinc finger and BTB domain containing 10
|
|
chr13_+_56804370
|
1.244
|
NM_008541
NM_001164041
|
Smad5
|
MAD homolog 5 (Drosophila)
|
|
chrX_+_12858142
|
1.205
|
NM_010028
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked
|
|
chr13_+_56805028
|
1.193
|
NM_001164042
|
Smad5
|
MAD homolog 5 (Drosophila)
|
|
chr15_+_73555336
|
1.189
|
NM_001166388
|
Ptp4a3
|
protein tyrosine phosphatase 4a3
|
|
chr2_-_59963796
|
1.166
|
NM_001001182
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr5_+_64203602
|
1.165
|
NM_029554
|
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene
|
|
chr11_+_87912647
|
1.136
|
NM_198013
|
Cuedc1
|
CUE domain containing 1
|
|
chr10_+_127814144
|
1.122
|
NM_172790
|
Ankrd52
|
ankyrin repeat domain 52
|
|
chr1_+_138521477
|
1.109
|
NM_001160251
NM_177643
|
Zfp281
|
zinc finger protein 281
|
|
chr5_+_32913539
|
1.097
|
NM_009535
|
Yes1
|
Yamaguchi sarcoma viral (v-yes) oncogene homolog 1
|
|
chr14_-_27353156
|
1.085
|
NM_032008
|
Slmap
|
sarcolemma associated protein
|
|
chr7_-_88837850
|
1.073
|
NM_001164087
|
Homer2
|
homer homolog 2 (Drosophila)
|
|
chr11_+_87983066
|
1.047
|
NM_001172099
|
Cuedc1
|
CUE domain containing 1
|
|
chr19_+_10257363
|
1.046
|
NM_146094
|
Fads1
|
fatty acid desaturase 1
|
|
chr10_+_42398721
|
1.043
|
NM_172416
|
Ostm1
|
osteopetrosis associated transmembrane protein 1
|
|
chr1_-_158604396
|
1.004
|
NM_023141
|
Tor3a
|
torsin family 3, member A
|
|
chr4_+_118781336
|
0.988
|
NM_011400
|
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr10_-_74980017
|
0.976
|
NM_175133
|
1110038D17Rik
|
RIKEN cDNA 1110038D17 gene
|
|
chr18_+_53335981
|
0.970
|
NM_026386
|
Snx2
|
sorting nexin 2
|
|
chr15_+_10106992
|
0.966
|
NM_011169
|
Prlr
|
prolactin receptor
|
|
chrX_-_160513980
|
0.948
|
NM_009173
|
Siah1b
|
seven in absentia 1B
|
|
chr14_+_75536138
|
0.947
|
NM_008879
|
Lcp1
|
lymphocyte cytosolic protein 1
|
|
chr5_+_93112460
|
0.901
|
NM_015756
|
Shroom3
|
shroom family member 3
|
|
chr6_+_142335733
|
0.870
|
NM_025872
|
Golt1b
|
golgi transport 1 homolog B (S. cerevisiae)
|
|
chrX_+_20265590
|
0.841
|
NM_011049
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr12_+_106270857
|
0.828
|
NM_028419
|
Glrx5
|
glutaredoxin 5 homolog (S. cerevisiae)
|
|
chr3_-_86978431
|
0.825
|
NM_001170985
NM_130867
|
Kirrel
|
kin of IRRE like (Drosophila)
|
|
chr15_-_96529998
|
0.822
|
NM_175121
|
Slc38a2
|
solute carrier family 38, member 2
|
|
chr13_+_112476313
|
0.808
|
NM_172593
|
Mier3
|
mesoderm induction early response 1, family member 3
|
|
chr7_-_91234908
|
0.799
|
NM_030728
|
9930013L23Rik
|
RIKEN cDNA 9930013L23 gene
|
|
chr10_-_44178474
|
0.798
|
NM_007548
|
Prdm1
|
PR domain containing 1, with ZNF domain
|
|
chr17_+_53706283
|
0.788
|
NM_020005
|
Kat2b
|
K(lysine) acetyltransferase 2B
|
|
chr4_+_128814102
|
0.767
|
NM_027402
|
Fndc5
|
fibronectin type III domain containing 5
|
|
chr9_+_77765170
|
0.732
|
NM_134255
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast)
|
|
chr3_-_144233110
|
0.730
|
NM_011828
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1
|
|
chr5_+_122608248
|
0.716
|
NM_013636
|
Ppp1cc
|
protein phosphatase 1, catalytic subunit, gamma isoform
|
|
chr3_+_32428382
|
0.711
|
NM_024200
|
Mfn1
|
mitofusin 1
|
|
chrX_+_96133099
|
0.704
|
NM_207633
|
Yipf6
|
Yip1 domain family, member 6
|
|
chr10_+_25128177
|
0.692
|
NM_013511
|
Epb4.1l2
|
erythrocyte protein band 4.1-like 2
|
|
chr12_+_70473032
|
0.676
|
NM_007481
|
Arf6
|
ADP-ribosylation factor 6
|
|
chrX_-_50622974
|
0.673
|
NM_001166365
NM_001166583
NM_030167
|
Fam122b
|
family with sequence similarity 122, member B
|
|
chr1_-_158649020
|
0.668
|
NM_145413
|
Fam20b
|
family with sequence similarity 20, member B
|
|
chr2_-_26300698
|
0.654
|
NM_153125
|
Sec16a
|
SEC16 homolog A (S. cerevisiae)
|
|
chrX_+_50265390
|
0.634
|
NM_027642
|
Phf6
|
PHD finger protein 6
|
|
chr8_+_109817355
|
0.634
|
NM_018823
NM_133957
|
Nfat5
|
nuclear factor of activated T-cells 5
|
|
chrX_+_69208336
|
0.625
|
NM_001081135
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr10_+_62383370
|
0.621
|
NM_175194
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16
|
|
chr3_-_122322556
|
0.612
|
NM_001114665
NM_153118
|
Fnbp1l
|
formin binding protein 1-like
|
|
chr8_+_108160437
|
0.608
|
NM_181322
|
Ctcf
|
CCCTC-binding factor
|
|
chr9_-_59334089
|
0.600
|
NM_019927
|
Arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila)
|
|
chr19_+_11986887
|
0.592
|
NM_172635
|
Patl1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr17_+_84911219
|
0.536
|
NM_177606
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr18_-_39646734
|
0.498
|
NM_008173
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1
|
|
chr4_+_135841981
|
0.485
|
NM_009557
|
Zfp46
|
zinc finger protein 46
|
|
chr11_-_98636538
|
0.469
|
NM_145434
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr5_+_122734363
|
0.456
|
NM_177242
|
Pptc7
|
PTC7 protein phosphatase homolog (S. cerevisiae)
|
|
chr5_-_36826896
|
0.439
|
NM_001170454
|
Tada2b
|
transcriptional adaptor 2B
|
|
chr2_-_106899807
|
0.400
|
NM_008045
|
Fshb
|
follicle stimulating hormone beta
|
|
chr6_-_33010151
|
0.376
|
NM_025336
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3
|
|
chr15_-_27955602
|
0.356
|
NM_001081302
|
Trio
|
triple functional domain (PTPRF interacting)
|
|
chr1_-_40847501
|
0.355
|
NM_172499
|
Mfsd9
|
major facilitator superfamily domain containing 9
|
|
chr6_-_119058206
|
0.353
|
NM_001159533
NM_001159534
NM_001159535
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr5_-_46247789
|
0.317
|
NM_172153
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like
|
|
chr2_-_156005873
|
0.300
|
NM_133242
|
Rbm39
|
RNA binding motif protein 39
|
|
chr17_+_53723450
|
0.285
|
NM_001190846
|
Kat2b
|
K(lysine) acetyltransferase 2B
|
|
chr7_+_87169699
|
0.278
|
NM_001145999
NM_001146000
|
Zfp710
|
zinc finger protein 710
|
|
chr5_-_144288812
|
0.258
|
NM_009007
|
Rac1
|
RAS-related C3 botulinum substrate 1
|
|
chr2_+_112219335
|
0.232
|
NM_024254
|
2410042D21Rik
|
RIKEN cDNA 2410042D21 gene
|