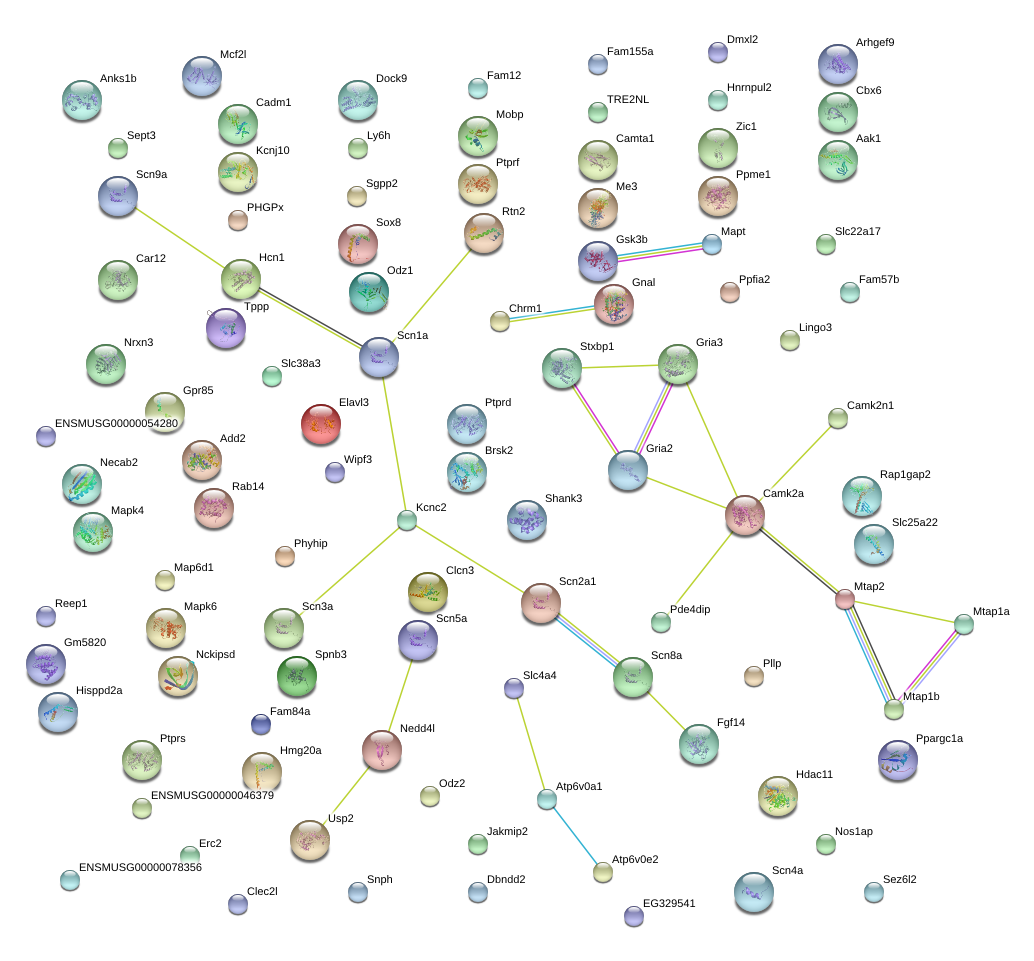
|
chr11_+_104092749
|
22.631
|
NM_001038609
NM_010838
|
Mapt
|
microtubule-associated protein tau
|
|
chr13_+_74146852
|
16.232
|
NM_182839
|
Tppp
|
tubulin polymerization promoting protein
|
|
chr2_-_151458256
|
15.784
|
NM_198214
|
Snph
|
syntaphilin
|
|
chr19_+_4711220
|
15.565
|
NM_021287
|
Spnb3
|
spectrin beta 3
|
|
chr8_+_12949417
|
14.471
|
NM_001159485
NM_001159486
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chr8_-_9771020
|
13.839
|
NM_173446
|
Fam155a
|
family with sequence similarity 155, member A
|
|
chr9_-_91260619
|
12.283
|
NM_009573
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr4_+_138011061
|
11.892
|
NM_025451
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr10_+_111708176
|
11.760
|
NM_001025581
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2
|
|
chr8_+_12915892
|
11.627
|
NM_178076
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chr1_+_66221851
|
11.590
|
NM_001039934
NM_008632
|
Mtap2
|
microtubule-associated protein 2
|
|
chr3_-_97628441
|
11.584
|
NM_001110163
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin)
|
|
chr6_+_71657791
|
11.396
|
NM_178608
|
Reep1
|
receptor accessory protein 1
|
|
chr9_-_107569654
|
11.244
|
NM_023805
|
Slc38a3
|
solute carrier family 38, member 3
|
|
chr9_-_54349409
|
11.216
|
NM_172771
|
Dmxl2
|
Dmx-like 2
|
|
chr18_-_43847367
|
11.056
|
NM_001163637
|
Jakmip2
|
janus kinase and microtubule interacting protein 2
|
|
chr7_-_107520359
|
11.053
|
NM_028292
|
Ppme1
|
protein phosphatase methylesterase 1
|
|
chrX_-_92361778
|
11.003
|
NM_001033329
|
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9
|
|
chr7_+_133968166
|
10.449
|
NM_001146347
|
Fam57b
|
family with sequence similarity 57, member B
|
|
chr3_-_97692613
|
10.257
|
NM_001039376
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin)
|
|
chr15_+_82105322
|
10.157
|
NM_011889
|
Sept3
|
septin 3
|
|
chr14_-_55531726
|
10.129
|
NM_021551
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17
|
|
chr2_+_164313584
|
10.022
|
NM_001048229
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr7_+_134094045
|
9.943
|
NM_144926
|
Sez6l2
|
seizure related 6 homolog like 2
|
|
chr18_+_67293403
|
9.814
|
NM_010307
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type
|
|
chr7_+_133966816
|
9.771
|
NM_029978
|
Fam57b
|
family with sequence similarity 57, member B
|
|
chr5_-_51945054
|
9.678
|
NM_008904
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr15_+_89330287
|
9.559
|
NM_021423
|
Shank3
|
SH3/ankyrin domain gene 3
|
|
chr2_+_65508501
|
9.541
|
NM_001099298
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1
|
|
chr2_+_164312376
|
9.332
|
NM_001048228
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr7_+_19876447
|
9.319
|
NM_001025364
|
Rtn2
|
reticulon 2 (Z-band associated protein)
|
|
chr15_-_75397601
|
9.265
|
NM_011837
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr9_+_120058842
|
9.112
|
NM_001039364
NM_001039365
NM_008614
|
Mobp
|
myelin-associated oligodendrocytic basic protein
|
|
chr2_-_65405546
|
8.987
|
NM_018732
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha
|
|
chr9_-_21856445
|
8.942
|
NM_010487
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr15_-_75397243
|
8.870
|
NM_001135688
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr1_+_78306890
|
8.781
|
NM_001004173
|
Sgpp2
|
sphingosine-1-phosphate phosphotase 2
|
|
chr9_+_43892984
|
8.668
|
NM_198091
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr1_+_174271338
|
8.457
|
NM_001039484
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr2_-_104250490
|
8.442
|
NM_001033347
|
D430041D05Rik
|
RIKEN cDNA D430041D05 gene
|
|
chr12_+_90191833
|
8.323
|
NM_172544
|
Nrxn3
|
neurexin III
|
|
chr2_-_66278870
|
8.316
|
NM_018733
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha
|
|
chr7_+_19868009
|
8.266
|
NM_013648
|
Rtn2
|
reticulon 2 (Z-band associated protein)
|
|
chr7_+_96781322
|
8.178
|
NM_181407
|
Me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr14_+_70857227
|
8.099
|
NM_145981
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein
|
|
chr18_-_74224541
|
7.982
|
NM_172632
|
Mapk4
|
mitogen-activated protein kinase 4
|
|
chr4_-_151235861
|
7.976
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr18_+_65183180
|
7.957
|
NM_031881
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like
|
|
chr15_-_75397030
|
7.908
|
NM_001135689
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr11_-_74403603
|
7.890
|
NM_001015046
|
Rap1gap2
|
RAP1 GTPase activating protein 2
|
|
chr10_+_89336253
|
7.835
|
NM_001128086
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_+_43877511
|
7.768
|
NM_016808
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr18_+_39152776
|
7.757
|
NM_175164
|
Arhgap26
|
Rho GTPase activating protein 26
|
|
chr2_+_164311639
|
7.745
|
NM_001048227
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr15_-_79635138
|
7.742
|
NM_030689
|
Nptxr
|
neuronal pentraxin receptor
|
|
chr9_+_108710676
|
7.715
|
NM_030729
|
Nckipsd
|
NCK interacting protein with SH3 domain
|
|
chr10_+_90038471
|
7.696
|
NM_001177397
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr17_-_25707599
|
7.511
|
NM_011447
|
Sox8
|
SRY-box containing gene 8
|
|
chr9_+_43875103
|
7.460
|
NM_198092
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr6_+_86799460
|
7.327
|
NM_001040106
NM_177762
|
Aak1
|
AP2 associated kinase 1
|
|
chr9_+_47338431
|
7.274
|
NM_001025600
NM_018770
NM_207675
NM_207676
|
Cadm1
|
cell adhesion molecule 1
|
|
chr2_+_121115337
|
7.249
|
NM_032393
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr2_-_121176719
|
7.190
|
NM_178795
|
Ppip5k1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr7_+_149135647
|
7.022
|
NM_029426
|
Brsk2
|
BR serine/threonine kinase 2
|
|
chr2_+_164311755
|
6.948
|
NM_026797
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr10_-_80306743
|
6.933
|
NM_001013758
|
Lingo3
|
leucine rich repeat and Ig domain containing 3
|
|
chr6_+_48487567
|
6.902
|
NM_133764
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2
|
|
chr15_-_84277488
|
6.874
|
NM_001163145
|
1810041L15Rik
|
RIKEN cDNA 1810041L15 gene
|
|
chr16_-_20241392
|
6.841
|
NM_198599
|
Map6d1
|
MAP6 domain containing 1
|
|
chrX_+_38754480
|
6.812
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr19_+_8738494
|
6.739
|
NM_001112697
NM_007698
|
Chrm1
|
cholinergic receptor, muscarinic 1, CNS
|
|
chr6_-_13789847
|
6.716
|
NM_145066
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr10_+_90039435
|
6.657
|
NM_001177396
NM_001177398
NM_181398
|
Ipw
Anks1b
|
imprinted gene in the Prader-Willi syndrome region
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr13_+_118391126
|
6.627
|
NM_010408
|
Hcn1
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 1
|
|
chr9_+_56266618
|
6.623
|
NM_025812
|
Hmg20a
|
high mobility group 20A
|
|
chr14_+_28435613
|
6.613
|
NM_177814
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr7_-_148623725
|
6.611
|
NM_026646
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22
|
|
chr8_+_121970569
|
6.573
|
NM_054095
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr10_+_105907394
|
6.563
|
NM_177373
|
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr2_+_121121189
|
6.531
|
NM_001173506
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr2_-_32702729
|
6.391
|
NM_009295
NM_001113569
|
Stxbp1
|
syntaxin binding protein 1
|
|
chr16_+_38089086
|
6.300
|
NM_019827
|
Gsk3b
|
glycogen synthase kinase 3 beta
|
|
chr6_+_38613031
|
6.277
|
NM_001101507
|
Clec2l
|
C-type lectin domain family, member L
|
|
chr6_+_91106773
|
6.252
|
NM_144919
|
Hdac11
|
histone deacetylase 11
|
|
chr8_-_63462080
|
6.228
|
NM_007711
NM_173873
NM_173874
|
Clcn3
|
chloride channel 3
|
|
chr8_-_97220127
|
6.204
|
NM_026385
|
Pllp
|
plasma membrane proteolipid
|
|
chr18_+_65047408
|
6.141
|
NM_001114386
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like
|
|
chr1_-_172519932
|
6.101
|
NM_001109985
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr4_-_76240202
|
6.040
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr11_-_36757743
|
6.022
|
NM_011856
|
Odz2
|
odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr7_+_133960398
|
6.016
|
NM_026884
|
Fam57b
|
family with sequence similarity 57, member B
|
|
chr12_-_14158843
|
5.970
|
NM_029007
|
Fam84a
|
family with sequence similarity 84, member A
|
|
chr18_+_67247984
|
5.951
|
NM_177137
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type
|
|
chr5_+_89316245
|
5.895
|
NM_001136260
NM_001197147
NM_018760
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4
|
|
chr6_+_54402876
|
5.858
|
NM_001167860
NM_001167861
|
Wipf3
|
WAS/WASL interacting protein family, member 3
|
|
chr14_-_125076242
|
5.856
|
NM_207667
|
Fgf14
|
fibroblast growth factor 14
|
|
chr5_-_33196860
|
5.783
|
NM_194340
|
C330019G07Rik
|
RIKEN cDNA C330019G07 gene
|
|
chr6_+_86028044
|
5.616
|
NM_013458
|
Add2
|
adducin 2 (beta)
|
|
chr7_-_148620437
|
5.589
|
NM_001177576
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22
|
|
chr14_-_122196929
|
5.579
|
NM_134074
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr13_-_100286521
|
5.563
|
NM_008634
|
Mtap1b
|
microtubule-associated protein 1B
|
|
chr6_+_124948454
|
5.542
|
NM_001145927
|
C530028O21Rik
|
RIKEN cDNA C530028O21 gene
|
|
chr11_+_100870759
|
5.525
|
NM_016920
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1
|
|
chr2_-_35056582
|
5.482
|
NM_026697
|
Rab14
|
RAB14, member RAS oncogene family
|
|
chr14_-_124591742
|
5.474
|
NM_010201
|
Fgf14
|
fibroblast growth factor 14
|
|
chr9_+_66561492
|
5.461
|
NM_178396
|
Car12
|
carbonic anyhydrase 12
|
|
chr6_+_124948627
|
5.451
|
NM_001145926
|
C530028O21Rik
|
RIKEN cDNA C530028O21 gene
|
|
chr18_+_61123204
|
5.444
|
NM_009792
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chrX_+_143397055
|
5.441
|
NM_008312
|
Htr2c
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr16_+_43247396
|
5.422
|
NM_019778
NM_181058
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr11_+_68912444
|
5.403
|
NM_011065
|
Per1
|
period homolog 1 (Drosophila)
|
|
chr2_+_156246644
|
5.364
|
NM_001003815
NM_013510
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr2_+_102498839
|
5.363
|
NM_001077514
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr1_+_89160921
|
5.349
|
NM_028889
|
Efhd1
|
EF hand domain containing 1
|
|
chr10_+_20760625
|
5.330
|
NM_001177776
|
Ahi1
|
Abelson helper integration site 1
|
|
chr3_+_95985065
|
5.216
|
NM_022030
|
Sv2a
|
synaptic vesicle glycoprotein 2 a
|
|
chr19_-_5085471
|
5.124
|
NM_001001885
|
Tmem151a
|
transmembrane protein 151A
|
|
chr6_-_112897259
|
5.120
|
NM_080448
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr11_+_89891661
|
5.115
|
NM_026433
|
Tmem100
|
transmembrane protein 100
|
|
chr7_+_116154373
|
5.107
|
NM_021885
|
Tub
|
tubby candidate gene
|
|
chr17_+_87153083
|
5.107
|
NM_010137
|
Epas1
|
endothelial PAS domain protein 1
|
|
chr9_+_21742717
|
5.088
|
NM_144935
|
BC018242
|
cDNA sequence BC018242
|
|
chr19_+_43514902
|
5.056
|
NM_031396
|
Cnnm1
|
cyclin M1
|
|
chr13_+_58907953
|
5.027
|
NM_008745
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr19_-_10315199
|
5.022
|
NM_001033481
|
Gm98
|
predicted gene 98
|
|
chr7_+_98239230
|
4.994
|
NM_011807
|
Dlg2
|
discs, large homolog 2 (Drosophila)
|
|
chr19_-_46401618
|
4.965
|
NM_028627
|
Psd
|
pleckstrin and Sec7 domain containing
|
|
chr11_+_77590766
|
4.963
|
NM_011586
|
Myo18a
|
myosin XVIIIA
|
|
chr1_+_89652645
|
4.956
|
NM_029846
|
Atg16l1
|
autophagy-related 16-like 1 (yeast)
|
|
chr2_-_154384574
|
4.939
|
NM_021546
|
Necab3
|
N-terminal EF-hand calcium binding protein 3
|
|
chr14_-_31736727
|
4.937
|
NM_008407
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3
|
|
chr4_-_75779379
|
4.936
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr8_+_97296066
|
4.927
|
NM_009142
|
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr14_-_88871202
|
4.914
|
NM_178685
|
Pcdh20
|
protocadherin 20
|
|
chr4_+_84851330
|
4.873
|
NM_019535
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr14_-_122097583
|
4.851
|
NM_001081039
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr5_-_118694869
|
4.808
|
NM_172998
NM_001109902
|
Rnft2
|
ring finger protein, transmembrane 2
|
|
chr4_-_151851574
|
4.801
|
NM_010598
|
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr16_-_46010510
|
4.781
|
NM_001134480
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr18_-_67708826
|
4.747
|
NM_176832
|
Spire1
|
spire homolog 1 (Drosophila)
|
|
chr11_+_49715292
|
4.692
|
NM_029004
|
Rasgef1c
|
RasGEF domain family, member 1C
|
|
chr17_-_24781872
|
4.670
|
NM_023449
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr16_-_34514112
|
4.635
|
NM_001164268
NM_177357
|
Kalrn
|
kalirin, RhoGEF kinase
|
|
chr3_+_105507516
|
4.632
|
NM_175398
|
6530418L21Rik
|
RIKEN cDNA 6530418L21 gene
|
|
chr12_+_89961317
|
4.616
|
NM_001198587
|
Nrxn3
|
neurexin III
|
|
chr5_-_139801315
|
4.612
|
NM_172723
|
Adap1
|
ArfGAP with dual PH domains 1
|
|
chr12_-_103943059
|
4.605
|
NM_172584
|
Itpk1
|
inositol 1,3,4-triphosphate 5/6 kinase
|
|
chr19_+_27291507
|
4.597
|
NM_001161420
NM_013703
|
Vldlr
|
very low density lipoprotein receptor
|
|
chr9_-_45714959
|
4.596
|
NM_178709
|
Rnf214
|
ring finger protein 214
|
|
chr18_-_62071451
|
4.562
|
NM_198649
|
Ablim3
|
actin binding LIM protein family, member 3
|
|
chr9_-_77392505
|
4.528
|
NM_001146048
|
Lrrc1
|
leucine rich repeat containing 1
|
|
chr9_+_64233432
|
4.505
|
NM_001134399
NM_172522
|
Megf11
|
multiple EGF-like-domains 11
|
|
chr1_+_66515003
|
4.482
|
NM_175510
|
Unc80
|
unc-80 homolog (C. elegans)
|
|
chr1_+_75139300
|
4.434
|
NM_170755
|
Fam134a
|
family with sequence similarity 134, member A
|
|
chr5_+_9266137
|
4.403
|
NM_172706
|
9330182L06Rik
|
RIKEN cDNA 9330182L06 gene
|
|
chr15_+_99532698
|
4.357
|
NM_031842
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr10_+_20672352
|
4.348
|
NM_026203
|
Ahi1
|
Abelson helper integration site 1
|
|
chr2_+_158201373
|
4.304
|
NM_175692
|
Snhg11
|
small nucleolar RNA host gene 11
|
|
chr6_-_85452851
|
4.294
|
NM_001001160
|
Fbxo41
|
F-box protein 41
|
|
chr2_-_92241357
|
4.284
|
NM_011162
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr19_+_10463540
|
4.280
|
NM_018801
NM_173067
NM_173068
|
Syt7
|
synaptotagmin VII
|
|
chr3_-_108218411
|
4.263
|
NM_001004177
NM_017392
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chrX_+_58962671
|
4.249
|
NM_001018087
|
Ldoc1
|
leucine zipper, down-regulated in cancer 1
|
|
chr8_-_113917651
|
4.230
|
NM_178086
|
Fa2h
|
fatty acid 2-hydroxylase
|
|
chr1_+_87791084
|
4.216
|
NM_022417
|
Itm2c
|
integral membrane protein 2C
|
|
chr2_+_181449669
|
4.215
|
NM_011012
|
Oprl1
|
opioid receptor-like 1
|
|
chr3_+_105255247
|
4.196
|
NM_001039347
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3
|
|
chr9_-_16182674
|
4.188
|
NM_001080814
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr8_+_77737842
|
4.175
|
NM_029182
|
Rasd2
|
RASD family, member 2
|
|
chr18_-_67712374
|
4.175
|
NM_194355
|
Spire1
|
spire homolog 1 (Drosophila)
|
|
chr18_-_62071494
|
4.150
|
NM_001164491
|
Ablim3
|
actin binding LIM protein family, member 3
|
|
chrX_-_7151185
|
4.135
|
NM_138605
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr1_+_60466462
|
4.119
|
NM_001198570
NM_001198571
NM_198127
|
Abi2
|
abl-interactor 2
|
|
chr2_-_91803660
|
4.104
|
NM_138306
|
Dgkz
|
diacylglycerol kinase zeta
|
|
chr4_-_148681792
|
4.104
|
NM_008441
NM_207682
|
Kif1b
|
kinesin family member 1B
|
|
chr3_-_31209216
|
4.078
|
NM_172861
|
Slc7a14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chrX_+_148613496
|
4.074
|
NM_001005475
|
Iqsec2
|
IQ motif and Sec7 domain 2
|
|
chr11_-_33893151
|
4.064
|
NM_001190886
|
Kcnip1
|
Kv channel-interacting protein 1
|
|
chr2_+_156301543
|
4.007
|
NM_001006664
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr8_-_97400173
|
3.986
|
NM_053246
|
Dok4
|
docking protein 4
|
|
chr7_-_88049882
|
3.931
|
NM_026523
|
Nmb
|
neuromedin B
|
|
chr14_-_122138082
|
3.919
|
NM_001128307
NM_001128308
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr11_-_72303293
|
3.913
|
NM_153060
|
Spns2
|
spinster homolog 2 (Drosophila)
|
|
chr19_-_14672472
|
3.879
|
NM_011600
|
Tle4
|
transducin-like enhancer of split 4, homolog of Drosophila E(spl)
|
|
chr2_-_91790504
|
3.870
|
NM_001166597
|
Dgkz
|
diacylglycerol kinase zeta
|
|
chr4_-_132895228
|
3.867
|
NM_199306
|
Wdtc1
|
WD and tetratricopeptide repeats 1
|
|
chr19_-_60937266
|
3.851
|
NM_053198
|
Sfxn4
|
sideroflexin 4
|
|
chr15_+_99548019
|
3.825
|
NM_010271
|
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr18_+_37120093
|
3.813
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr18_+_37127284
|
3.790
|
NM_007767
|
Pcdha6
|
protocadherin alpha 6
|
|
chr12_-_88607158
|
3.735
|
NM_001142580
NM_001142581
|
Vipar
|
VPS33B interacting protein, apical-basolateral polarity regulator
|
|
chr19_-_42506122
|
3.707
|
NM_145123
|
Crtac1
|
cartilage acidic protein 1
|
|
chr19_-_5118229
|
3.672
|
NM_008451
|
Klc2
|
kinesin light chain 2
|
|
chr19_-_4175478
|
3.671
|
NM_134148
|
Carns1
|
carnosine synthase 1
|
|
chr11_+_75345164
|
3.637
|
NM_173388
|
Slc43a2
|
solute carrier family 43, member 2
|
|
chr6_-_92517359
|
3.632
|
NM_001134461
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr18_+_37098858
|
3.625
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr6_-_57485419
|
3.595
|
NM_175523
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing)
|
|
chr2_-_25174662
|
3.587
|
NM_001177656
NM_001177657
NM_008169
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1)
|
|
chr2_-_130468507
|
3.581
|
NM_197945
|
Prosapip1
|
ProSAPiP1 protein
|