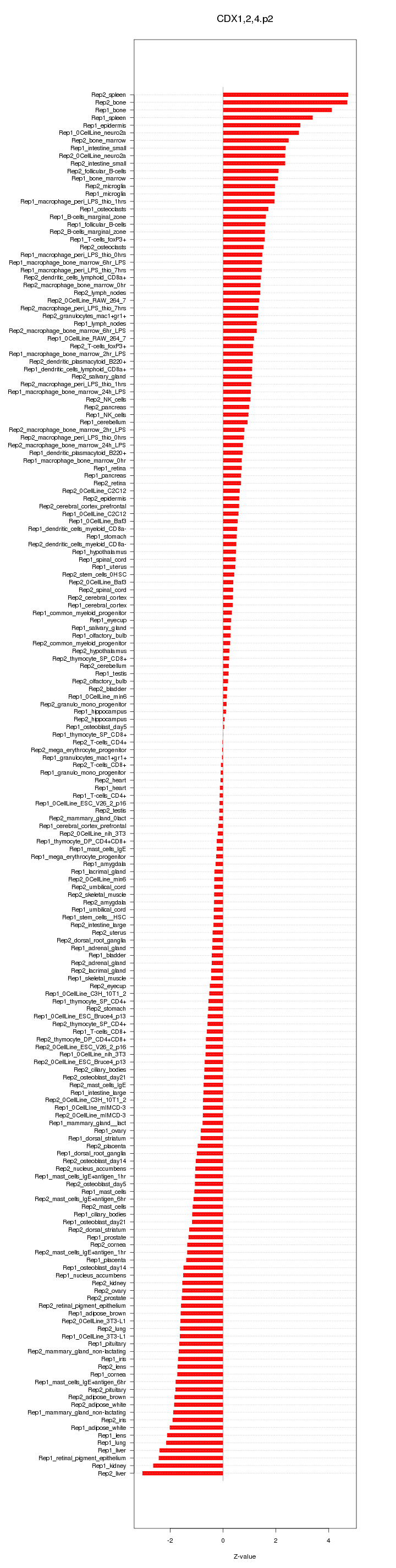
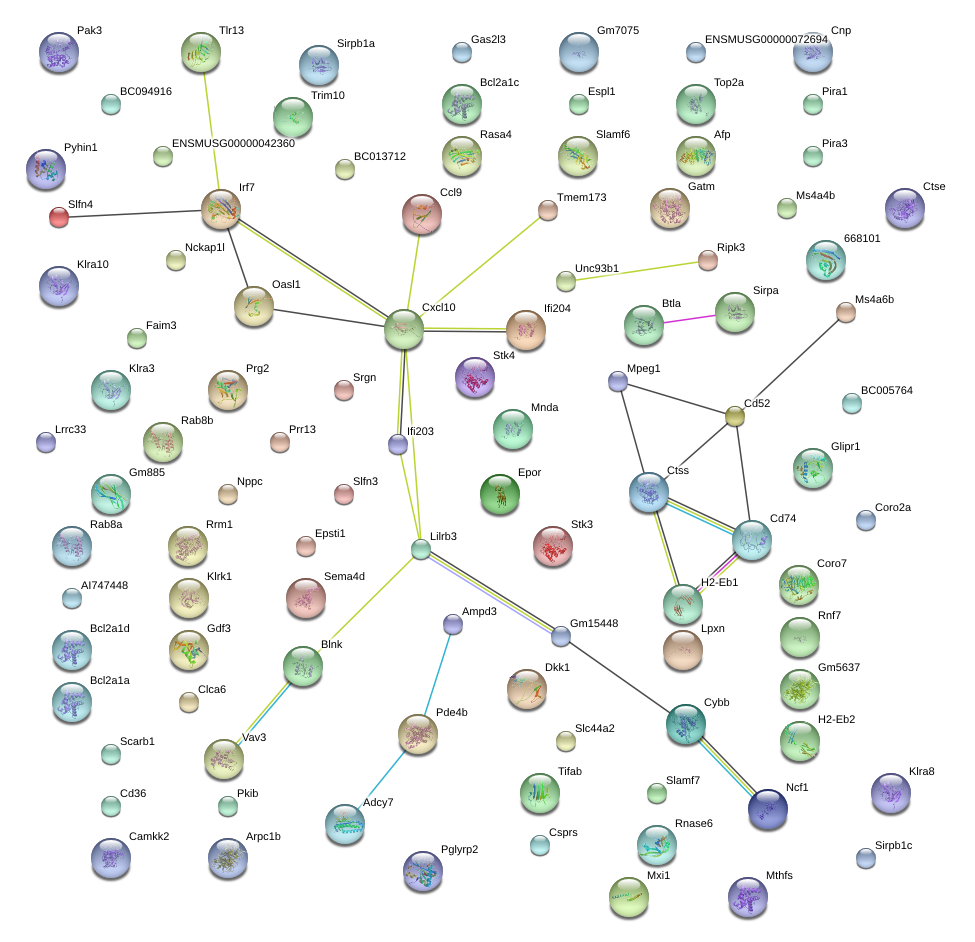
|
chr13_-_56280161
|
17.499
|
NM_001168615
NM_145976
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B
|
|
chr18_+_60963495
|
16.334
|
NM_001042605
NM_010545
|
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated)
|
|
chr11_+_82988687
|
15.703
|
NM_011410
|
Slfn4
|
schlafen 4
|
|
chr5_+_115373272
|
15.598
|
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr3_+_109376824
|
15.375
|
NM_146139
|
Vav3
|
vav 3 oncogene
|
|
chr3_+_95330700
|
15.184
|
NM_021281
|
Ctss
|
cathepsin S
|
|
chr9_-_88626434
|
14.809
|
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr11_-_98859189
|
14.320
|
|
Top2a
|
topoisomerase (DNA) II alpha
|
|
chr3_-_15426426
|
14.289
|
NM_001002898
|
Sirpb1a
|
signal-regulatory protein beta 1A
|
|
chr17_+_34442837
|
13.978
|
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr3_-_15748418
|
13.531
|
NM_001173459
|
LOC100038947
Sirpb1a
|
signal-regulatory protein beta 1-like
signal-regulatory protein beta 1A
|
|
chr19_-_41069024
|
13.511
|
NM_008528
|
Blnk
|
B-cell linker
|
|
chr19_+_12535536
|
13.043
|
|
Mpeg1
|
macrophage expressed gene 1
|
|
chr19_+_12873115
|
12.694
|
|
Lpxn
|
leupaxin
|
|
chr14_+_51748742
|
12.155
|
NM_030098
|
Rnase6
|
ribonuclease, RNase A family, 6
|
|
chr8_+_90812964
|
12.092
|
NM_001037723
|
Adcy7
|
adenylate cyclase 7
|
|
chr1_+_175560938
|
11.711
|
NM_175026
|
Pyhin1
|
pyrin and HIN domain family, member 1
|
|
chr19_+_53403902
|
11.656
|
NM_010847
|
Mxi1
|
Max interacting protein 1
|
|
chr10_-_88875526
|
11.302
|
|
Gas2l3
|
growth arrest-specific 2 like 3
|
|
chrX_+_103338609
|
11.191
|
NM_205820
|
Tlr13
|
toll-like receptor 13
|
|
chr1_+_132762248
|
11.164
|
|
Faim3
|
Fas apoptotic inhibitory molecule 3
|
|
chr6_-_70067577
|
11.138
|
|
Gm10883
|
predicted gene 10883
|
|
chr5_+_115373246
|
11.111
|
NM_145209
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr6_-_129572590
|
11.108
|
NM_033078
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr19_+_12535268
|
11.096
|
NM_010821
|
Mpeg1
|
macrophage expressed gene 1
|
|
chr17_+_34442808
|
10.690
|
NM_010382
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr2_+_163899842
|
10.576
|
NM_021420
|
Stk4
|
serine/threonine kinase 4
|
|
chr5_+_90919768
|
10.545
|
|
Afp
|
alpha fetoprotein
|
|
chr15_+_102289600
|
10.431
|
NM_001170911
NM_025385
|
Prr13
|
proline rich 13
|
|
chr14_-_56407622
|
10.329
|
NM_001164107
NM_001164108
NM_019955
|
Ripk3
|
receptor-interacting serine-threonine kinase 3
|
|
chr2_-_122420914
|
10.056
|
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr16_-_4679664
|
9.979
|
NM_030205
|
Coro7
|
coronin 7
|
|
chr15_+_103284200
|
9.914
|
NM_153505
|
Nckap1l
|
NCK associated protein 1 like
|
|
chr18_-_35900005
|
9.528
|
NM_028261
|
Tmem173
|
transmembrane protein 173
|
|
chr11_+_87569075
|
9.331
|
|
A430104N18Rik
|
RIKEN cDNA A430104N18 gene
|
|
chr17_+_37006518
|
9.274
|
NM_011280
|
Trim10
|
tripartite motif-containing 10
|
|
chr1_+_132762353
|
9.109
|
NM_026976
|
Faim3
|
Fas apoptotic inhibitory molecule 3
|
|
chr19_+_12873063
|
9.020
|
NM_134152
|
Lpxn
|
leupaxin
|
|
chr1_+_173847645
|
8.977
|
NM_030710
|
Slamf6
|
SLAM family member 6
|
|
chr7_-_25873560
|
8.946
|
|
|
|
|
chr5_-_92776160
|
8.757
|
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chrX_-_9030859
|
8.591
|
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr10_-_61960613
|
8.580
|
|
Srgn
|
serglycin
|
|
chr19_+_3935169
|
8.536
|
NM_001161428
NM_019449
|
Unc93b1
|
unc-93 homolog B1 (C. elegans)
|
|
chr5_-_134705423
|
8.466
|
NM_010876
|
Ncf1
|
neutrophil cytosolic factor 1
|
|
chr5_+_90919758
|
7.928
|
|
Afp
|
alpha fetoprotein
|
|
chr11_-_83388330
|
7.593
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr4_-_46579311
|
7.553
|
NM_178893
|
Coro2a
|
coronin, actin binding protein 2A
|
|
chr19_-_30623890
|
7.493
|
NM_010051
|
Dkk1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr7_+_109614267
|
7.478
|
|
Rrm1
|
ribonucleotide reductase M1
|
|
chr2_+_84820615
|
7.376
|
NM_008920
|
Prg2
|
proteoglycan 2, bone marrow
|
|
chr10_-_111434265
|
7.369
|
NM_028608
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr7_+_117916117
|
7.317
|
NM_009667
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr17_-_32557916
|
7.257
|
NM_021319
|
Pglyrp2
|
peptidoglycan recognition protein 2
|
|
chr15_+_102126686
|
7.222
|
NM_001014976
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr10_-_111434231
|
7.168
|
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr14_+_78304045
|
7.108
|
NM_029495
NM_178825
|
Epsti1
|
epithelial stromal interaction 1 (breast)
|
|
chr5_+_136559785
|
7.059
|
NM_001039103
NM_133914
|
Rasa4
|
RAS p21 protein activator 4
|
|
chr16_+_45224449
|
6.860
|
NM_001037719
NM_177584
|
Btla
|
B and T lymphocyte associated
|
|
chr4_-_132352272
|
6.851
|
NM_001033308
|
BC013712
|
cDNA sequence BC013712
|
|
chr11_+_83037063
|
6.702
|
|
AI662270
|
expressed sequence AI662270
|
|
chr4_-_133650937
|
6.572
|
NM_013706
|
Cd52
|
CD52 antigen
|
|
chr13_-_51829951
|
6.466
|
|
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr19_-_41068625
|
6.463
|
|
Blnk
|
B-cell linker
|
|
chr9_-_21767872
|
6.426
|
NM_010149
|
Epor
|
erythropoietin receptor
|
|
chr7_+_4225744
|
6.341
|
NM_011093
NM_008848
|
Pira6
|
paired-Ig-like receptor A6
|
|
chr9_+_89094190
|
6.335
|
|
Bcl2a1a
|
B-cell leukemia/lymphoma 2 related protein A1a
|
|
chr16_-_32165509
|
6.170
|
NM_146069
|
Lrrc33
|
leucine rich repeat containing 33
|
|
chr11_+_100442602
|
6.147
|
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr5_+_145888588
|
6.123
|
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B
|
|
chr6_-_122560002
|
6.050
|
|
Gdf3
|
growth differentiation factor 3
|
|
chr5_-_17355533
|
6.036
|
NM_001159556
NM_001159557
|
Cd36
|
CD36 antigen
|
|
chr7_-_3796619
|
6.023
|
|
Pira1
Pira2
Lilrb3
|
paired-Ig-like receptor A1
paired-Ig-like receptor A2
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
|
|
chr5_-_123183537
|
5.918
|
|
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr1_+_133535033
|
5.917
|
|
Ctse
|
cathepsin E
|
|
chr2_+_129442079
|
5.915
|
|
Sirpa
|
signal-regulatory protein alpha
|
|
chr11_+_106612525
|
5.904
|
NM_001033435
|
Gm885
|
predicted gene 885
|
|
chr19_+_11518042
|
5.848
|
NM_021718
|
Ms4a4b
|
membrane-spanning 4-domains, subfamily A, member 4B
|
|
chr19_+_11592938
|
5.754
|
NM_027209
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B
|
|
chr7_-_148452299
|
5.694
|
NM_016850
|
Irf7
|
interferon regulatory factor 7
|
|
chr3_-_144595486
|
5.631
|
NM_001033199
|
AI747448
|
expressed sequence AI747448
|
|
chr10_+_57370786
|
5.620
|
NM_001039050
NM_001190402
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific
|
|
chr9_+_21142267
|
5.612
|
NM_152808
|
Slc44a2
|
solute carrier family 44, member 2
|
|
chr4_+_102242664
|
5.472
|
NM_001177980
|
Pde4b
|
phosphodiesterase 4B, cAMP specific
|
|
chr5_-_17341534
|
5.453
|
|
Cd36
|
CD36 antigen
|
|
chr6_-_130287518
|
5.402
|
NM_010648
|
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3
|
|
chr9_-_66767436
|
5.356
|
NM_173413
|
Rab8b
|
RAB8B, member RAS oncogene family
|
|
chr1_-_175805812
|
5.348
|
|
Mndal
|
myeloid nuclear differentiation antigen like
|
|
chr1_-_173579057
|
5.258
|
|
Slamf7
|
SLAM family member 7
|
|
chr15_+_102126735
|
5.249
|
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr9_-_96379093
|
5.232
|
|
Rnf7
|
ring finger protein 7
|
|
chr1_-_167638156
|
5.184
|
NM_001038846
NM_178593
|
Rcsd1
|
RCSD domain containing 1
|
|
chr7_-_113358823
|
5.183
|
NM_001039160
NM_029000
|
Gvin1
|
GTPase, very large interferon inducible 1
|
|
chr7_-_113102453
|
5.183
|
NM_001039160
NM_029000
|
Gvin1
|
GTPase, very large interferon inducible 1
|
|
chr7_+_134439804
|
5.167
|
NM_008400
|
Itgal
|
integrin alpha L
|
|
chr7_-_21288567
|
5.165
|
NM_001167155
NM_001167154
|
Gm4144
Gm8693
|
predicted gene 4144
predicted gene 8693
|
|
chr5_+_86500770
|
5.162
|
NM_019992
|
Stap1
|
signal transducing adaptor family member 1
|
|
chr1_+_173612132
|
5.151
|
|
Cd48
|
CD48 antigen
|
|
chr12_+_109843975
|
5.142
|
NM_001163395
|
Evl
|
Ena-vasodilator stimulated phosphoprotein
|
|
chr16_-_75909440
|
5.134
|
NM_023380
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1
|
|
chr14_+_48092244
|
5.113
|
|
Fbxo34
|
F-box protein 34
|
|
chr6_-_122560022
|
5.098
|
NM_008108
|
Gdf3
|
growth differentiation factor 3
|
|
chr14_+_48092305
|
5.080
|
|
Fbxo34
|
F-box protein 34
|
|
chr17_+_35189798
|
4.899
|
|
|
|
|
chr6_+_128837604
|
4.838
|
NM_020257
|
Clec2i
|
C-type lectin domain family 2, member i
|
|
chr5_-_88956788
|
4.808
|
NM_152839
|
Igj
|
immunoglobulin joining chain
|
|
chr11_+_62413710
|
4.783
|
|
|
|
|
chr18_-_36926866
|
4.748
|
|
|
|
|
chr12_+_82049290
|
4.716
|
|
Srsf5
|
serine/arginine-rich splicing factor 5
|
|
chr11_-_83462722
|
4.714
|
NM_011337
|
Ccl3
|
chemokine (C-C motif) ligand 3
|
|
chr2_-_27118325
|
4.696
|
|
Vav2
|
vav 2 oncogene
|
|
chr7_-_23477335
|
4.677
|
NM_001167155
NM_001167154
|
Gm4144
Gm8693
|
predicted gene 4144
predicted gene 8693
|
|
chr12_-_115170216
|
4.654
|
|
Ighg
|
Immunoglobulin heavy chain (gamma polypeptide)
|
|
chr9_-_14185685
|
4.624
|
NM_028013
|
Endod1
|
endonuclease domain containing 1
|
|
chr8_+_119983664
|
4.624
|
|
|
|
|
chr1_+_173612173
|
4.575
|
NM_007649
|
Cd48
|
CD48 antigen
|
|
chr12_-_27129497
|
4.543
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr13_-_67500685
|
4.536
|
NM_001080941
|
Zfp429
|
zinc finger protein 429
|
|
chr19_+_36453556
|
4.511
|
NM_029508
|
Pcgf5
|
polycomb group ring finger 5
|
|
chr7_-_22813915
|
4.500
|
NM_001166839
NM_001167169
NM_001167168
NM_001166746
|
Gm4172
Gm4179
Gm4220
Vmn1r111
|
predicted gene 4172
predicted gene 4179
predicted gene 4220
vomeronasal 1 receptor 111
|
|
chr9_+_7272513
|
4.460
|
NM_008607
|
Mmp13
|
matrix metallopeptidase 13
|
|
chr13_+_24706474
|
4.436
|
NM_178658
|
Fam65b
|
family with sequence similarity 65, member B
|
|
chr14_-_103379725
|
4.377
|
|
Kctd12
|
potassium channel tetramerisation domain containing 12
|
|
chr11_+_60955985
|
4.365
|
|
Tnfrsf13b
|
tumor necrosis factor receptor superfamily, member 13b
|
|
chr3_-_112959644
|
4.346
|
NM_001190403
NM_001190404
|
Amy2b
|
amylase 2b
|
|
chr11_-_34647110
|
4.320
|
NM_027411
|
Ccdc99
|
coiled-coil domain containing 99
|
|
chr4_+_135728188
|
4.311
|
NM_177733
|
E2f2
|
E2F transcription factor 2
|
|
chr7_+_29225955
|
4.307
|
NM_001039192
|
Gmfg
|
glia maturation factor, gamma
|
|
chr10_+_79457716
|
4.277
|
|
Cnn2
|
calponin 2
|
|
chr2_+_24242134
|
4.272
|
|
Psd4
|
pleckstrin and Sec7 domain containing 4
|
|
chr10_+_79348991
|
4.266
|
NM_015779
|
Elane
|
elastase, neutrophil expressed
|
|
chr1_-_173582969
|
4.240
|
NM_144539
|
Slamf7
|
SLAM family member 7
|
|
chr12_-_80084464
|
4.206
|
NM_001170401
|
Tmem229b
|
transmembrane protein 229B
|
|
chr14_-_20796255
|
4.206
|
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr1_-_55044032
|
4.197
|
|
Sf3b1
|
splicing factor 3b, subunit 1
|
|
chr1_-_167638124
|
4.183
|
|
Rcsd1
|
RCSD domain containing 1
|
|
chr1_-_192994532
|
4.172
|
|
Atf3
|
activating transcription factor 3
|
|
chr1_+_109407998
|
4.138
|
NM_001174170
NM_011111
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2
|
|
chr5_-_17341512
|
4.135
|
|
Cd36
|
CD36 antigen
|
|
chr1_-_53813191
|
4.120
|
|
|
|
|
chr11_-_115375672
|
4.104
|
NM_008258
|
Hn1
|
hematological and neurological expressed sequence 1
|
|
chr7_-_3820753
|
4.085
|
|
Gm10693
|
predicted pseudogene 10693
|
|
chr3_-_113094211
|
4.069
|
NM_001160150
NM_001160151
NM_001160152
NM_001042711
|
Amy2a4
Amy2a3
Amy2a2
Amy2a5
|
amylase 2a4
amylase 2a3
amylase 2a2
amylase 2a5
|
|
chr5_+_34868420
|
4.054
|
NM_011893
|
Sh3bp2
|
SH3-domain binding protein 2
|
|
chr17_+_34375847
|
4.052
|
NM_010389
|
H2-Ob
|
histocompatibility 2, O region beta locus
|
|
chr9_+_124016886
|
4.046
|
NM_009915
|
Ccr2
|
chemokine (C-C motif) receptor 2
|
|
chr6_-_129422702
|
4.025
|
NM_020008
|
Clec7a
|
C-type lectin domain family 7, member a
|
|
chr3_-_113126814
|
4.022
|
NM_001160150
NM_001160151
NM_001160152
NM_001042711
|
Amy2a4
Amy2a3
Amy2a2
Amy2a5
|
amylase 2a4
amylase 2a3
amylase 2a2
amylase 2a5
|
|
chr5_-_17394564
|
4.021
|
|
Cd36
|
CD36 antigen
|
|
chr10_+_57351786
|
3.996
|
NM_001039051
NM_008863
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific
|
|
chr11_-_98862163
|
3.991
|
|
Top2a
|
topoisomerase (DNA) II alpha
|
|
chr3_+_145813137
|
3.990
|
NM_001005846
|
Mcoln2
|
mucolipin 2
|
|
chr17_+_48594225
|
3.974
|
NM_152823
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like
|
|
chr7_-_21488234
|
3.960
|
NM_001166839
NM_001167169
NM_001167168
NM_001166746
|
Gm4172
Gm4179
Gm4220
Vmn1r111
|
predicted gene 4172
predicted gene 4179
predicted gene 4220
vomeronasal 1 receptor 111
|
|
chr7_-_24036700
|
3.960
|
NM_001166839
NM_001167169
NM_001167168
NM_001166746
|
Gm4172
Gm4179
Gm4220
Vmn1r111
|
predicted gene 4172
predicted gene 4179
predicted gene 4220
vomeronasal 1 receptor 111
|
|
chr1_+_60803231
|
3.938
|
NM_007642
|
Cd28
|
CD28 antigen
|
|
chr7_-_3796651
|
3.923
|
NM_011089
|
Pira2
|
paired-Ig-like receptor A2
|
|
chr5_-_105411580
|
3.920
|
NM_030239
|
Abcg3
|
ATP-binding cassette, sub-family G (WHITE), member 3
|
|
chr14_-_20796411
|
3.896
|
NM_001038637
NM_010315
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr4_+_44313764
|
3.885
|
NM_010790
|
Melk
|
maternal embryonic leucine zipper kinase
|
|
chr8_+_34763709
|
3.871
|
NM_010344
|
Gsr
|
glutathione reductase
|
|
chr10_+_66648458
|
3.867
|
|
Jmjd1c
|
jumonji domain containing 1C
|
|
chr4_+_44313865
|
3.854
|
|
Melk
|
maternal embryonic leucine zipper kinase
|
|
chr3_+_145784635
|
3.850
|
NM_134160
|
Mcoln3
|
mucolipin 3
|
|
chr8_+_74121544
|
3.849
|
NM_001166213
|
Fam129c
|
family with sequence similarity 129, member C
|
|
chr7_+_29222465
|
3.827
|
NM_022024
|
Gmfg
|
glia maturation factor, gamma
|
|
chr1_-_64167818
|
3.825
|
|
Klf7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr11_-_83391984
|
3.819
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr10_-_61970481
|
3.817
|
NM_011157
|
Srgn
|
serglycin
|
|
chr17_-_32557807
|
3.804
|
|
Pglyrp2
|
peptidoglycan recognition protein 2
|
|
chr3_+_95465860
|
3.790
|
|
Mcl1
|
myeloid cell leukemia sequence 1
|
|
chr7_+_28954717
|
3.789
|
NM_007991
|
Fbl
|
fibrillarin
|
|
chr11_+_100442831
|
3.787
|
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr2_-_148263617
|
3.743
|
|
Cd93
|
CD93 antigen
|
|
chr15_-_82074765
|
3.728
|
NM_001080158
|
Cenpm
|
centromere protein M
|
|
chr7_+_22057197
|
3.710
|
NM_001166740
|
Vmn1r125
|
vomeronasal 1 receptor 125
|
|
chr6_+_116123984
|
3.706
|
|
Gm8203
|
predicted pseudogene 8203
|
|
chr5_+_34014563
|
3.682
|
|
|
|
|
chr16_+_36184297
|
3.665
|
NM_001082547
|
Gm5483
|
predicted gene 5483
|
|
chr9_-_123771243
|
3.647
|
NM_011798
|
Xcr1
|
chemokine (C motif) receptor 1
|
|
chr8_+_72332573
|
3.631
|
NM_198101
|
Gmip
|
Gem-interacting protein
|
|
chr7_+_23656242
|
3.623
|
NM_001166724
|
Vmn1r160
|
vomeronasal 1 receptor 160
|
|
chr6_+_83065466
|
3.620
|
NM_020619
|
Mogs
|
mannosyl-oligosaccharide glucosidase
|
|
chr14_-_20796199
|
3.610
|
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr13_-_101086551
|
3.608
|
NM_010871
|
Naip6
|
NLR family, apoptosis inhibitory protein 6
|
|
chr4_-_46417054
|
3.605
|
NM_053149
|
Hemgn
|
hemogen
|
|
chr5_+_114580397
|
3.602
|
NM_001040691
|
Ung
|
uracil DNA glycosylase
|
|
chr5_+_150076605
|
3.586
|
NM_009663
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein
|
|
chrX_+_83115611
|
3.575
|
NM_001163539
|
5430427O19Rik
|
RIKEN cDNA 5430427O19 gene
|
|
chr7_-_22331108
|
3.569
|
NM_001166839
NM_001167169
NM_001167168
NM_001166746
|
Gm4172
Gm4179
Gm4220
Vmn1r111
|
predicted gene 4172
predicted gene 4179
predicted gene 4220
vomeronasal 1 receptor 111
|
|
chr3_-_113159420
|
3.560
|
NM_001160150
NM_001160151
NM_001160152
NM_001042711
|
Amy2a4
Amy2a3
Amy2a2
Amy2a5
|
amylase 2a4
amylase 2a3
amylase 2a2
amylase 2a5
|
|
chr11_-_34647016
|
3.544
|
|
Ccdc99
|
coiled-coil domain containing 99
|
|
chr7_+_63185457
|
3.521
|
|
Cyfip1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr8_+_85689250
|
3.515
|
NM_001111304
NM_027758
|
Tbc1d9
|
TBC1 domain family, member 9
|
|
chr14_-_122314994
|
3.507
|
NM_182806
|
Gpr18
|
G protein-coupled receptor 18
|
|
chr7_-_21697365
|
3.500
|
NM_001167151
NM_001167150
NM_001166742
|
Gm4498
Gm8453
Vmn1r118
|
predicted gene 4498
predicted gene 8453
vomeronasal 1 receptor 118
|
|
chr14_-_52780511
|
3.452
|
|
Supt16h
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr17_+_34282215
|
3.450
|
NM_010388
|
H2-DMb1
H2-DMb2
|
histocompatibility 2, class II, locus Mb1
histocompatibility 2, class II, locus Mb2
|
|
chr9_-_82826139
|
3.431
|
|
Phip
|
pleckstrin homology domain interacting protein
|
|
chr7_-_23628623
|
3.425
|
NM_001166758
|
Vmn1r159
|
vomeronasal 1 receptor 159
|