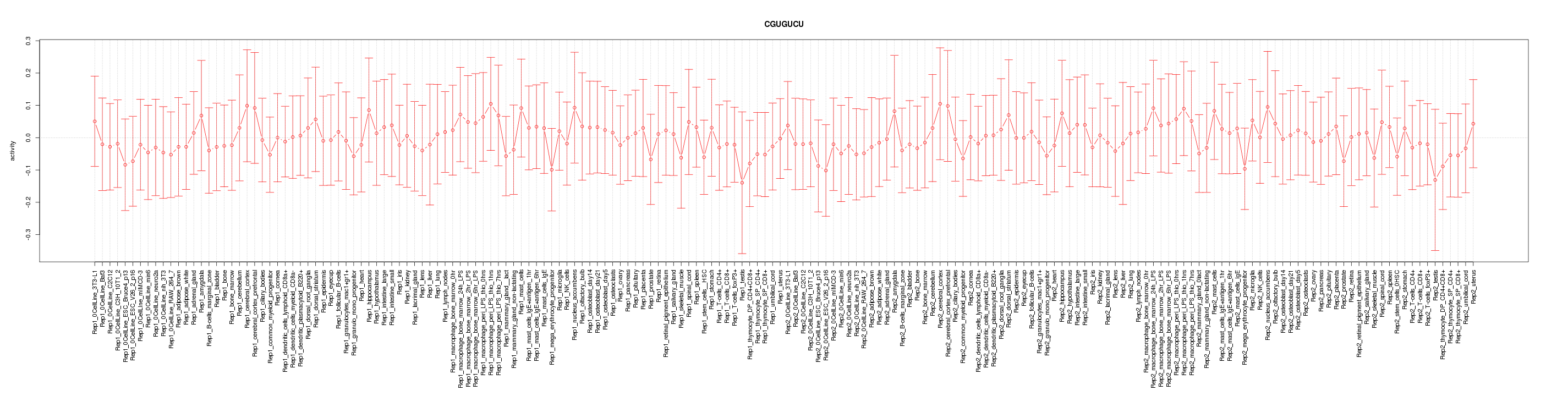
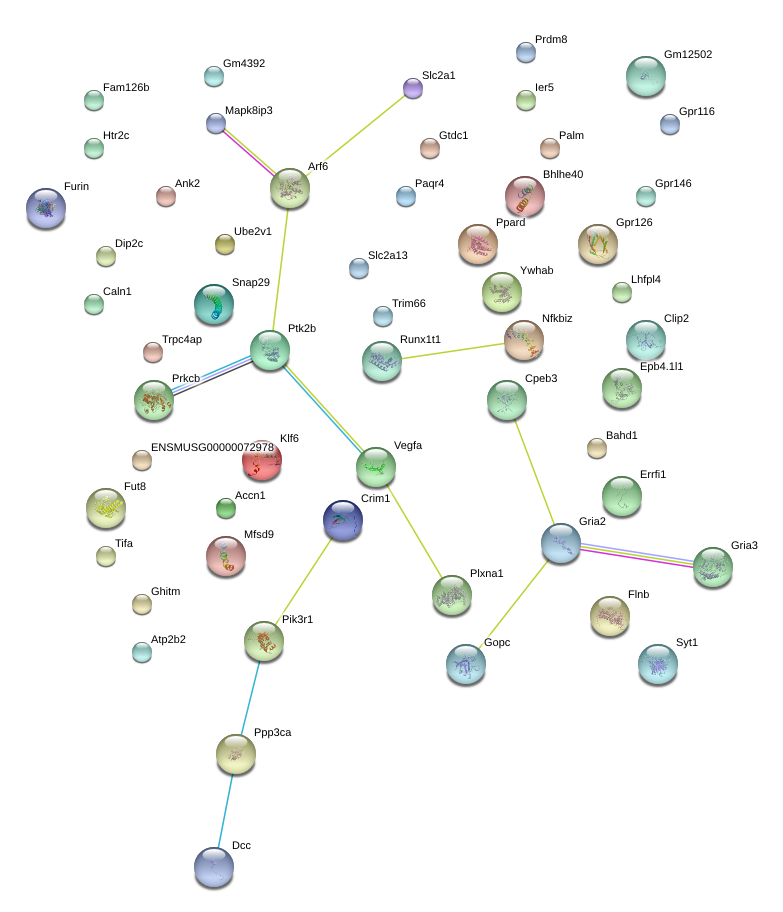
|
chr7_+_129432585
|
2.341
|
NM_008855
|
Prkcb
|
protein kinase C, beta
|
|
chr3_-_126646303
|
2.135
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr6_+_108610606
|
2.133
|
NM_011498
|
Bhlhe40
|
basic helix-loop-helix family, member e40
|
|
chr14_-_66832383
|
2.026
|
NM_001162366
|
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta
|
|
chr5_+_130845319
|
1.864
|
NM_181045
|
Caln1
|
calneuron 1
|
|
chr2_+_156246644
|
1.835
|
NM_001003815
NM_013510
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr4_+_118781336
|
1.811
|
NM_011400
|
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr14_-_66899764
|
1.761
|
NM_001162365
NM_172498
|
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta
|
|
chr6_-_113841356
|
1.721
|
NM_001036684
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr15_-_91403685
|
1.673
|
NM_001033633
|
Slc2a13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr3_-_126701367
|
1.672
|
NM_178655
|
Ank2
|
ankyrin 2, brain
|
|
chr6_-_113991945
|
1.649
|
NM_009723
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr5_+_130924661
|
1.644
|
NM_021371
|
Caln1
|
calneuron 1
|
|
chr3_+_136333733
|
1.633
|
NM_008913
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform
|
|
chr3_+_127492716
|
1.613
|
NM_145133
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain
|
|
chr10_-_108447674
|
1.276
|
NM_009306
|
Syt1
|
synaptotagmin I
|
|
chrX_+_38754480
|
1.257
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr2_+_156301543
|
1.153
|
NM_001006664
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr17_-_25073897
|
1.073
|
NM_001163447
NM_001163448
NM_001163449
NM_001163450
NM_001163451
NM_001163453
NM_013931
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr7_-_116651647
|
1.051
|
NM_001170912
NM_181853
|
Trim66
|
tripartite motif-containing 66
|
|
chr17_-_46162111
|
0.951
|
NM_001110266
NM_001110267
NM_001110268
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr13_+_9275741
|
0.860
|
NM_001081426
|
Dip2c
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr11_-_80966253
|
0.853
|
NM_007384
|
Accn1
|
amiloride-sensitive cation channel 1, neuronal (degenerin)
|
|
chr14_-_37948251
|
0.835
|
NM_078478
|
Ghitm
|
growth hormone inducible transmembrane protein
|
|
chr11_-_81781897
|
0.832
|
NM_001034013
|
Accn1
|
amiloride-sensitive cation channel 1, neuronal (degenerin)
|
|
chrX_+_143397055
|
0.821
|
NM_008312
|
Htr2c
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr10_-_52101880
|
0.775
|
NM_053187
|
Gopc
|
golgi associated PDZ and coiled-coil motif containing
|
|
chr13_+_5860701
|
0.769
|
NM_011803
|
Klf6
|
Kruppel-like factor 6
|
|
chr1_-_156946739
|
0.756
|
NM_010500
|
Ier5
|
immediate early response 5
|
|
chr5_-_135028197
|
0.752
|
NM_001039162
NM_009990
|
Clip2
|
CAP-GLY domain containing linker protein 2
|
|
chr17_-_23877115
|
0.720
|
NM_023824
|
Paqr4
|
progestin and adipoQ receptor family member IV
|
|
chr16_-_55838747
|
0.716
|
NM_001159395
NM_030612
|
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr18_-_72510670
|
0.670
|
NM_007831
|
Dcc
|
deleted in colorectal carcinoma
|
|
chr4_+_13711928
|
0.636
|
NM_009822
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_98609872
|
0.594
|
NM_029947
|
Prdm8
|
PR domain containing 8
|
|
chr6_-_89312545
|
0.574
|
NM_008881
|
Plxna1
|
plexin A1
|
|
chr4_+_13670425
|
0.536
|
NM_001111026
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_+_150229178
|
0.513
|
NM_133753
|
Errfi1
|
ERBB receptor feedback inhibitor 1
|
|
chr7_-_116637251
|
0.505
|
NM_001170913
|
Trim66
|
tripartite motif-containing 66
|
|
chr17_-_46169322
|
0.487
|
NM_001025250
NM_001025257
NM_009505
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr10_+_79256298
|
0.484
|
NM_001161747
NM_023128
|
Palm
|
paralemmin
|
|
chr16_-_55822217
|
0.474
|
NM_001159394
|
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr1_-_40847501
|
0.473
|
NM_172499
|
Mfsd9
|
major facilitator superfamily domain containing 9
|
|
chr2_+_118727248
|
0.464
|
NM_001045523
|
Bahd1
|
bromo adjacent homology domain containing 1
|
|
chr2_-_155518064
|
0.436
|
NM_001163452
NM_019828
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr16_+_17406078
|
0.322
|
NM_023348
|
Snap29
|
synaptosomal-associated protein 29
|
|
chr1_-_58642994
|
0.274
|
NM_172513
|
Fam126b
|
family with sequence similarity 126, member B
|
|
chr10_-_14264800
|
0.256
|
NM_001002268
|
Gpr126
|
G protein-coupled receptor 126
|
|
chr12_+_70473032
|
0.253
|
NM_007481
|
Arf6
|
ADP-ribosylation factor 6
|
|
chr14_+_8650470
|
0.230
|
NM_134080
|
Flnb
|
filamin, beta
|
|
chr2_+_163820920
|
0.172
|
NM_018753
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr17_+_28369642
|
0.152
|
NM_011145
|
Ppard
|
peroxisome proliferator activator receptor delta
|
|
chr5_+_139865857
|
0.151
|
NM_001038703
|
Gpr146
|
G protein-coupled receptor 146
|
|
chr12_+_78339999
|
0.137
|
NM_016893
|
Fut8
|
fucosyltransferase 8
|
|
chr17_+_78599556
|
0.134
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr4_+_13678443
|
0.128
|
NM_001111027
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr19_-_37281762
|
0.122
|
NM_198300
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr5_+_139856534
|
0.118
|
NM_030258
|
Gpr146
|
G protein-coupled receptor 146
|
|
chr2_-_44717141
|
0.110
|
NM_172662
|
Gtdc1
|
glycosyltransferase-like domain containing 1
|
|
chr2_-_167457476
|
0.096
|
NM_023230
|
Ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr13_-_102462584
|
0.095
|
NM_001024955
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)
|
|
chr6_-_113145321
|
0.077
|
NM_177763
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4
|
|
chr7_-_87550315
|
0.038
|
NM_001081454
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr17_+_43526389
|
0.020
|
NM_001081178
|
Gpr116
|
G protein-coupled receptor 116
|