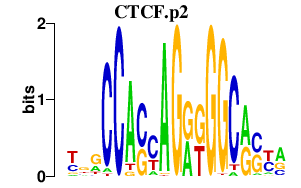
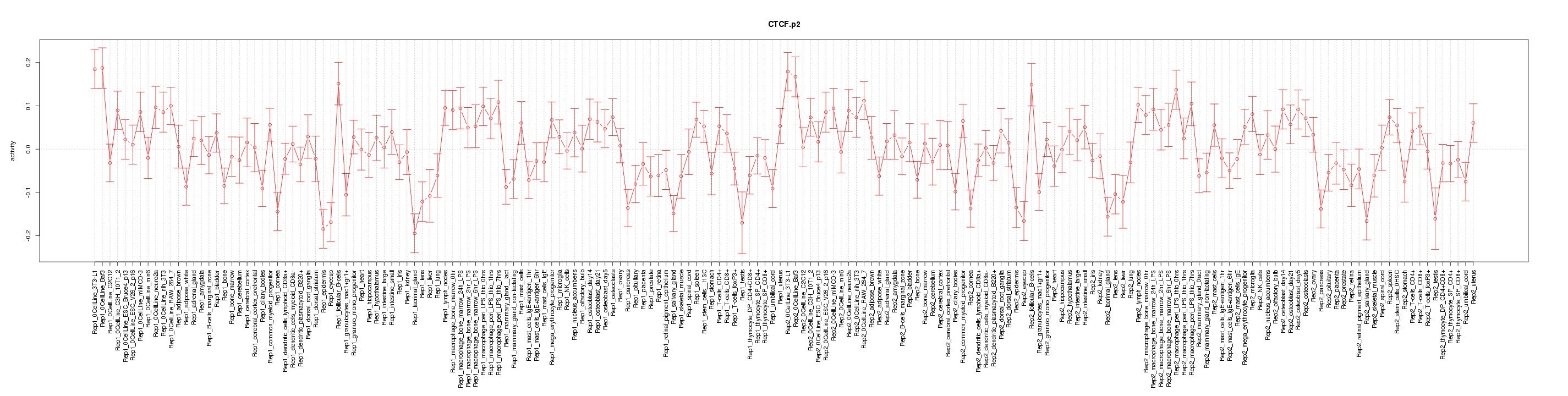
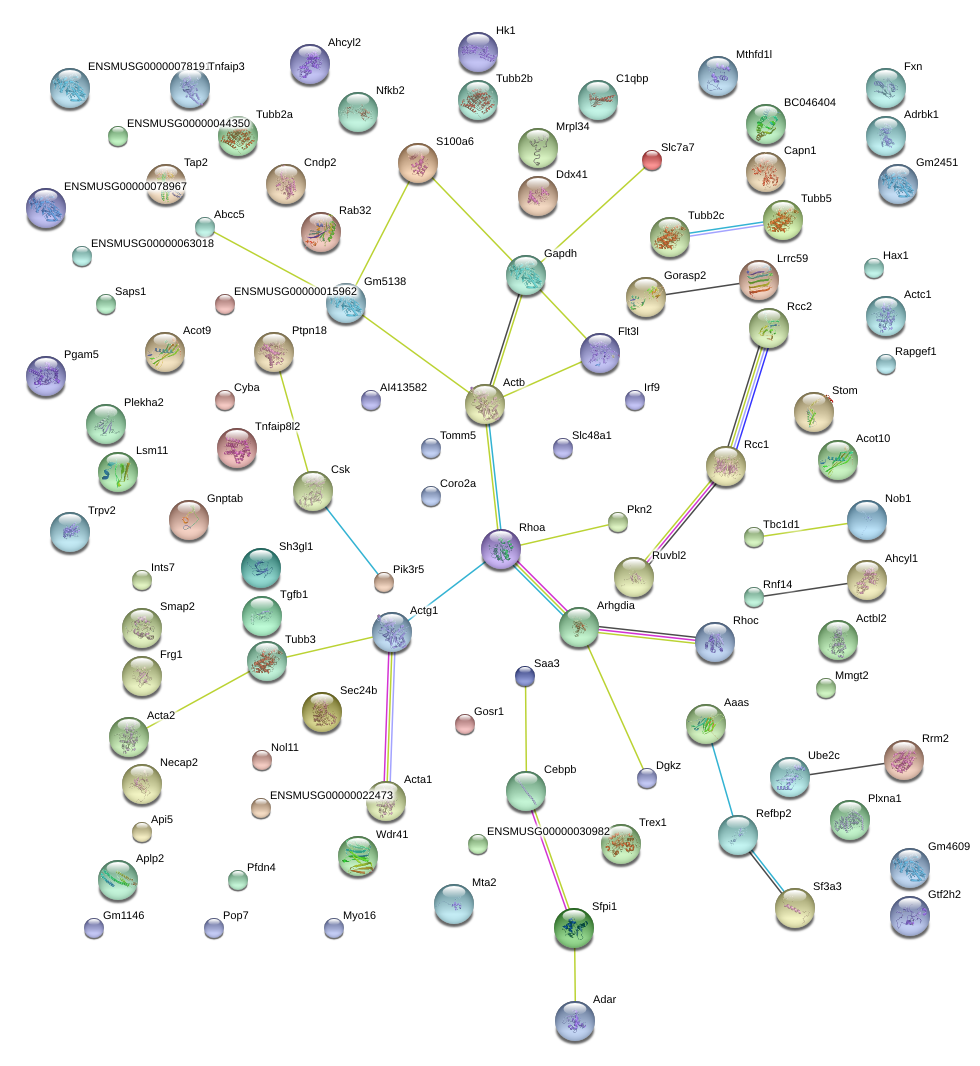
|
chr8_-_26212588
|
16.271
|
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2
|
|
chr7_+_26471968
|
14.473
|
NM_011577
|
Tgfb1
|
transforming growth factor, beta 1
|
|
chr11_+_68245608
|
12.976
|
NM_177320
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5, p101
|
|
chr3_-_107499129
|
12.399
|
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1
|
|
chr17_-_35973192
|
12.361
|
|
Tubb5
|
tubulin, beta 5
|
|
chr11_+_62389367
|
12.223
|
|
Trpv2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr11_-_120441643
|
11.938
|
|
Arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr11_-_107050573
|
11.831
|
NM_001161329
NM_133702
|
Nol11
|
nucleolar protein 11
|
|
chr7_-_53970378
|
11.749
|
|
Saa3
|
serum amyloid A 3
|
|
chr11_-_45758413
|
10.601
|
NM_028185
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11
|
|
chr2_+_170336729
|
10.563
|
NM_001110152
|
Pfdn4
|
prefoldin 4
|
|
chr3_-_94946256
|
10.340
|
NM_027206
|
Tnfaip8l2
|
tumor necrosis factor, alpha-induced protein 8-like 2
|
|
chr10_-_6373309
|
10.331
|
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr2_+_90936845
|
10.226
|
NM_011355
|
Sfpi1
|
SFFV proviral integration 1
|
|
chr10_-_6373388
|
10.170
|
NM_001170785
NM_001170786
NM_172308
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr17_-_35972880
|
10.079
|
|
Tubb5
|
tubulin, beta 5
|
|
chr19_+_46379216
|
9.715
|
NM_001177369
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2, p49/p100
|
|
chr3_-_129763586
|
9.561
|
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae)
|
|
chr2_+_170336930
|
9.236
|
|
Pfdn4
|
prefoldin 4
|
|
chr19_-_6015778
|
8.830
|
NM_001110504
|
Capn1
|
calpain 1
|
|
chr17_+_34341417
|
8.821
|
NM_011530
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr4_+_140257340
|
8.735
|
NM_173867
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr17_+_34341467
|
8.701
|
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr10_-_10277663
|
8.697
|
|
Rab32
|
RAB32, member RAS oncogene family
|
|
chr5_-_110698882
|
8.690
|
|
Pgam5
|
phosphoglycerate mutase family member 5
|
|
chr5_-_110698874
|
8.389
|
|
Pgam5
|
phosphoglycerate mutase family member 5
|
|
chr7_-_52689833
|
8.380
|
NM_011304
|
Ruvbl2
|
RuvB-like protein 2
|
|
chr10_-_10277885
|
8.290
|
NM_026405
|
Rab32
|
RAB32, member RAS oncogene family
|
|
chr6_-_89312545
|
8.146
|
NM_008881
|
Plxna1
|
plexin A1
|
|
chr6_-_125112509
|
8.141
|
|
Gapdh
Gm5523
Gm10481
|
glyceraldehyde-3-phosphate dehydrogenase
glyceraldehyde-3-phosphate dehydrogenase pseudogene
glyceraldehyde-3-phosphate dehydrogenase pseudogene
|
|
chr10_-_61738537
|
8.065
|
|
Hk1
|
hexokinase 1
|
|
chr19_-_4305954
|
8.053
|
NM_130863
|
Adrbk1
|
adrenergic receptor kinase, beta 1
|
|
chr11_+_94491055
|
8.027
|
NM_133807
|
Lrrc59
|
leucine rich repeat containing 59
|
|
chr5_-_110698814
|
7.962
|
|
Pgam5
|
phosphoglycerate mutase family member 5
|
|
chr13_-_55637866
|
7.924
|
NM_134059
|
Ddx41
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41
|
|
chr8_-_124956759
|
7.868
|
NM_007806
|
Cyba
|
cytochrome b-245, alpha polypeptide
|
|
chr8_+_73988823
|
7.721
|
NM_053162
|
Mrpl34
|
mitochondrial ribosomal protein L34
|
|
chr11_-_76577056
|
7.566
|
NM_016810
|
Gosr1
|
golgi SNAP receptor complex member 1
|
|
chr11_+_62462273
|
7.564
|
|
Mmgt2
|
membrane magnesium transporter 2
|
|
chr9_-_108962219
|
7.560
|
NM_011637
|
Trex1
|
three prime repair exonuclease 1
|
|
chr11_+_62462314
|
7.467
|
|
Mmgt2
|
membrane magnesium transporter 2
|
|
chr5_-_110698902
|
7.450
|
NM_001163538
NM_028273
|
Pgam5
|
phosphoglycerate mutase family member 5
|
|
chr3_+_90417072
|
7.416
|
|
S100a6
|
S100 calcium binding protein A6 (calcyclin)
|
|
chr2_+_164595389
|
7.410
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr2_+_70499545
|
7.376
|
NM_027352
|
Gorasp2
|
golgi reassembly stacking protein 2
|
|
chr17_-_27702625
|
7.149
|
NM_001002895
|
AI413582
|
expressed sequence AI413582
|
|
chr7_+_125883784
|
7.100
|
NM_027815
|
9030624J02Rik
|
RIKEN cDNA 9030624J02 gene
|
|
chr12_+_25393083
|
7.053
|
NM_009104
|
Rrm2
|
ribonucleotide reductase M2
|
|
chr11_-_62462024
|
7.021
|
NM_198861
|
BC046404
|
cDNA sequence BC046404
|
|
chr2_+_164595415
|
6.962
|
NM_026785
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr3_+_90416815
|
6.933
|
NM_011313
|
S100a6
|
S100 calcium binding protein A6 (calcyclin)
|
|
chr7_-_111613624
|
6.884
|
NM_009099
|
Trim30a
|
tripartite motif-containing 30A
|
|
chr9_-_57492978
|
6.780
|
NM_007783
|
Csk
|
c-src tyrosine kinase
|
|
chr14_-_77436363
|
6.693
|
NM_172488
|
9030625A04Rik
|
RIKEN cDNA 9030625A04 gene
|
|
chr10_+_87841876
|
6.659
|
NM_001004164
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits
|
|
chr2_+_167514414
|
6.571
|
NM_009883
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr1_+_34529567
|
6.571
|
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18
|
|
chr13_-_101262544
|
6.523
|
NM_022011
|
Gtf2h2
|
general transcription factor II H, polypeptide 2
|
|
chr11_-_76577079
|
6.484
|
|
Gosr1
|
golgi SNAP receptor complex member 1
|
|
chr8_-_26212203
|
6.474
|
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2
|
|
chr19_-_4306221
|
6.433
|
|
Adrbk1
|
adrenergic receptor kinase, beta 1
|
|
chr7_-_111656285
|
6.414
|
NM_001167828
NM_199146
|
Trim30a
Trim30d
|
tripartite motif-containing 30A
tripartite motif-containing 30D
|
|
chr1_+_173433608
|
6.378
|
NM_019484
|
Refbp2
|
RNA and export factor binding protein 2
|
|
chr3_-_89802501
|
6.363
|
NM_011826
|
Hax1
|
HCLS1 associated X-1
|
|
chr15_+_97614772
|
6.312
|
NM_026353
|
Slc48a1
|
solute carrier family 48 (heme transporter), member 1
|
|
chr15_-_89204248
|
6.249
|
NM_001111288
|
Sco2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr4_-_140634151
|
6.232
|
NM_025383
|
Necap2
|
NECAP endocytosis associated 2
|
|
chr2_+_29475402
|
6.186
|
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr15_-_102181072
|
6.082
|
NM_153416
|
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia
|
|
chr8_-_42502362
|
6.075
|
NM_013522
|
Frg1
|
FSHD region gene 1
|
|
chr9_-_57493393
|
6.016
|
|
Csk
|
c-src tyrosine kinase
|
|
chr3_+_89534639
|
5.995
|
NM_001146296
NM_019655
|
Adar
|
adenosine deaminase, RNA-specific
|
|
chr3_+_104591955
|
5.995
|
|
Rhoc
|
ras homolog gene family, member C
|
|
chr10_-_18735154
|
5.966
|
NM_009397
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr17_-_35089399
|
5.819
|
|
1110038B12Rik
|
RIKEN cDNA 1110038B12 gene
|
|
chr18_+_38456315
|
5.776
|
|
Rnf14
|
ring finger protein 14
|
|
chr1_+_173433762
|
5.751
|
|
Refbp2
|
RNA and export factor binding protein 2
|
|
chr17_-_56175896
|
5.688
|
NM_013664
|
Sh3gl1
|
SH3-domain GRB2-like 1
|
|
chr18_+_38456364
|
5.646
|
NM_001164621
|
Rnf14
|
ring finger protein 14
|
|
chr2_+_167514489
|
5.596
|
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr3_-_142544883
|
5.581
|
NM_178654
|
Pkn2
|
protein kinase N2
|
|
chr3_+_104591946
|
5.557
|
NM_007484
|
Rhoc
|
ras homolog gene family, member C
|
|
chr5_+_64550623
|
5.525
|
|
Tbc1d1
|
TBC1 domain family, member 1
|
|
chr18_-_84849981
|
5.522
|
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr14_-_55036515
|
5.504
|
NM_011405
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr8_-_109948891
|
5.494
|
NM_026277
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae)
|
|
chr2_-_35192443
|
5.478
|
NM_013515
|
Stom
|
stomatin
|
|
chr3_-_129763648
|
5.466
|
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae)
|
|
chr19_-_24355054
|
5.460
|
NM_008044
|
Fxn
|
frataxin
|
|
chr14_+_56222821
|
5.432
|
NM_001159417
NM_008394
|
Irf9
|
interferon regulatory factor 9
|
|
chr19_+_9026020
|
5.431
|
|
Mta2
|
metastasis-associated gene family, member 2
|
|
chrX_-_6998125
|
5.405
|
|
2010204K13Rik
|
RIKEN cDNA 2010204K13 gene
|
|
chr3_-_107499460
|
5.365
|
NM_145542
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1
|
|
chr2_-_91803660
|
5.319
|
NM_138306
|
Dgkz
|
diacylglycerol kinase zeta
|
|
chr3_-_129763732
|
5.305
|
NM_207209
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae)
|
|
chr7_-_52391419
|
5.283
|
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand
|
|
chrX_-_7000037
|
5.255
|
|
2010204K13Rik
|
RIKEN cDNA 2010204K13 gene
|
|
chr4_+_124392019
|
5.245
|
NM_029157
|
Sf3a3
|
splicing factor 3a, subunit 3
|
|
chr11_-_70796432
|
5.219
|
|
C1qbp
|
complement component 1, q subcomponent binding protein
|
|
chr18_+_38456280
|
5.213
|
NM_001164622
NM_020012
|
Rnf14
|
ring finger protein 14
|
|
chr4_-_46614726
|
5.212
|
NM_001164804
|
Coro2a
|
coronin, actin binding protein 2A
|
|
chr7_-_4610179
|
5.190
|
|
Ppp6r1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr2_+_29475234
|
5.157
|
NM_001039086
NM_001039087
NM_054050
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr2_-_94278303
|
5.109
|
NM_007466
|
Api5
|
apoptosis inhibitor 5
|
|
chr7_-_4610551
|
5.102
|
NM_172894
|
Ppp6r1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr1_+_193399604
|
5.101
|
NM_178632
|
Ints7
|
integrator complex subunit 7
|
|
chr13_+_95746298
|
5.078
|
NM_172590
|
Wdr41
|
WD repeat domain 41
|
|
chr5_-_137943570
|
5.032
|
NM_028753
|
Pop7
|
processing of precursor 7, ribonuclease P family, (S. cerevisiae)
|
|
chr9_-_31019412
|
5.026
|
|
Aplp2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr16_-_20426357
|
5.022
|
NM_013790
NM_176839
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr2_+_167514460
|
5.003
|
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr4_-_131901576
|
4.947
|
NM_001197082
NM_133878
|
Rcc1
|
regulator of chromosome condensation 1
|
|
chr11_-_70796471
|
4.859
|
NM_007573
|
C1qbp
|
complement component 1, q subcomponent binding protein
|
|
chr4_-_45120942
|
4.846
|
NM_001099675
NM_001134646
|
Tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast)
|
|
chrX_+_151696991
|
4.843
|
NM_019736
|
Acot9
|
acyl-CoA thioesterase 9
|
|
chr14_+_56223001
|
4.825
|
|
Irf9
|
interferon regulatory factor 9
|
|
chrX_+_155970637
|
4.742
|
|
A830080D01Rik
|
RIKEN cDNA A830080D01 gene
|
|
chr4_-_120642197
|
4.734
|
|
Smap2
|
stromal membrane-associated GTPase-activating protein 2
|
|
chrX_+_155970593
|
4.725
|
NM_001033472
|
A830080D01Rik
|
RIKEN cDNA A830080D01 gene
|
|
chr5_+_64551449
|
4.693
|
NM_019636
|
Tbc1d1
|
TBC1 domain family, member 1
|
|
chr5_-_143667004
|
4.666
|
|
Actb
|
actin, beta
|
|
chr11_+_5688463
|
4.628
|
NM_001146308
NM_001146309
NM_013810
|
Dbnl
|
drebrin-like
|
|
chr12_+_8320219
|
4.626
|
NM_021429
|
Hs1bp3
|
HCLS1 binding protein 3
|
|
chr5_-_137943329
|
4.606
|
|
Pop7
|
processing of precursor 7, ribonuclease P family, (S. cerevisiae)
|
|
chr5_+_115956809
|
4.602
|
|
Pxn
|
paxillin
|
|
chr1_+_155596555
|
4.592
|
NM_011882
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent)
|
|
chr7_+_16894925
|
4.577
|
NM_133234
|
Bbc3
|
BCL2 binding component 3
|
|
chr6_+_108733075
|
4.567
|
|
Arl8b
|
ADP-ribosylation factor-like 8B
|
|
chr6_-_3349415
|
4.549
|
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr19_-_6141100
|
4.530
|
NM_019722
|
Arl2
|
ADP-ribosylation factor-like 2
|
|
chr10_+_24589791
|
4.527
|
NM_001166416
NM_027347
|
Med23
|
mediator complex subunit 23
|
|
chr17_-_34027488
|
4.438
|
|
Kifc1
|
kinesin family member C1
|
|
chr2_+_151319915
|
4.426
|
NM_198326
|
Nsfl1c
|
NSFL1 (p97) cofactor (p47)
|
|
chr14_+_56223363
|
4.420
|
NM_001159418
|
Irf9
|
interferon regulatory factor 9
|
|
chr8_-_20020246
|
4.408
|
|
2610005L07Rik
A430108E01Rik
6820431F20Rik
|
cadherin 11 pseudogene
RIKEN cDNA A430108E01 gene
cadherin 11 pseudogene
|
|
chr6_+_108733041
|
4.408
|
NM_026011
|
Arl8b
|
ADP-ribosylation factor-like 8B
|
|
chr10_-_126467848
|
4.399
|
NM_025537
|
Tsfm
|
Ts translation elongation factor, mitochondrial
|
|
chr11_-_76391228
|
4.378
|
NM_198894
|
Abr
|
active BCR-related gene
|
|
chr5_+_116009516
|
4.369
|
|
Rplp0
|
ribosomal protein, large, P0
|
|
chr12_+_101202708
|
4.363
|
NM_001164426
NM_001164427
NM_146037
|
Kcnk13
|
potassium channel, subfamily K, member 13
|
|
chr1_-_53841997
|
4.337
|
NM_133810
|
Stk17b
|
serine/threonine kinase 17b (apoptosis-inducing)
|
|
chr13_-_45097365
|
4.299
|
NM_025772
|
Dtnbp1
|
dystrobrevin binding protein 1
|
|
chr19_+_5878465
|
4.278
|
NM_134154
|
Slc25a45
|
solute carrier family 25, member 45
|
|
chr14_-_22668093
|
4.257
|
NM_026965
|
Comtd1
|
catechol-O-methyltransferase domain containing 1
|
|
chr2_-_118199046
|
4.247
|
NM_201367
|
Gpr176
|
G protein-coupled receptor 176
|
|
chr1_-_87633103
|
4.245
|
NM_177055
|
A630001G21Rik
|
RIKEN cDNA A630001G21 gene
|
|
chr14_+_66915852
|
4.244
|
NM_029979
|
Trim35
|
tripartite motif-containing 35
|
|
chr5_+_115685856
|
4.190
|
NM_026398
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae)
|
|
chr3_+_14641726
|
4.185
|
NM_024495
|
Car13
|
carbonic anhydrase 13
|
|
chr17_+_35068258
|
4.180
|
|
Neu1
|
neuraminidase 1
|
|
chr15_-_98568883
|
4.176
|
|
|
|
|
chr1_+_82836029
|
4.147
|
NM_010472
|
Agfg1
|
ArfGAP with FG repeats 1
|
|
chr16_+_16303044
|
4.124
|
NM_198246
|
Yars2
|
tyrosyl-tRNA synthetase 2 (mitochondrial)
|
|
chr7_-_52391756
|
4.110
|
NM_013520
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand
|
|
chr8_+_23303277
|
4.105
|
NM_027338
|
Vps36
|
vacuolar protein sorting 36 (yeast)
|
|
chr2_+_170321927
|
4.058
|
NM_001013369
|
Pfdn4
|
prefoldin 4
|
|
chr2_-_94278124
|
4.056
|
|
Api5
|
apoptosis inhibitor 5
|
|
chr5_-_140212063
|
4.023
|
NM_174850
|
Micall2
|
MICAL-like 2
|
|
chr10_-_117147757
|
3.962
|
NM_010786
|
Mdm2
|
transformed mouse 3T3 cell double minute 2
|
|
chr15_+_76175485
|
3.956
|
|
|
|
|
chr8_-_72149443
|
3.955
|
NM_172754
|
Zfp868
|
zinc finger protein 868
|
|
chr14_+_66915867
|
3.927
|
|
Trim35
|
tripartite motif-containing 35
|
|
chr2_-_114000981
|
3.874
|
|
Aqr
|
aquarius
|
|
chr10_-_117147722
|
3.845
|
|
Mdm2
|
transformed mouse 3T3 cell double minute 2
|
|
chr7_+_134944106
|
3.828
|
NM_001040684
NM_133943
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr1_+_120218417
|
3.789
|
NM_026472
|
Mki67ip
|
Mki67 (FHA domain) interacting nucleolar phosphoprotein
|
|
chr11_+_83286587
|
3.781
|
NM_027427
|
Taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor
|
|
chr16_-_20425991
|
3.779
|
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr2_+_28368603
|
3.730
|
NM_001145834
NM_009058
|
Ralgds
|
ral guanine nucleotide dissociation stimulator
|
|
chr15_+_97794629
|
3.727
|
NM_201359
|
Tmem106c
|
transmembrane protein 106C
|
|
chr11_+_67466129
|
3.727
|
|
Gas7
|
growth arrest specific 7
|
|
chr11_+_67346499
|
3.725
|
NM_008088
|
Gas7
|
growth arrest specific 7
|
|
chr19_+_46130978
|
3.722
|
NM_001081214
|
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1
|
|
chr11_+_78141923
|
3.718
|
NM_201406
|
Pigs
|
phosphatidylinositol glycan anchor biosynthesis, class S
|
|
chr5_-_5749270
|
3.716
|
NM_027399
|
Steap1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chr7_-_52391649
|
3.713
|
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand
|
|
chr11_+_88835235
|
3.704
|
NM_016706
|
Coil
|
coilin
|
|
chr8_-_27104573
|
3.677
|
NM_029037
|
4930444A02Rik
|
RIKEN cDNA 4930444A02 gene
|
|
chr1_+_120218430
|
3.661
|
|
Mki67ip
|
Mki67 (FHA domain) interacting nucleolar phosphoprotein
|
|
chr3_-_105490320
|
3.642
|
NM_017397
|
Ddx20
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20
|
|
chr8_-_72149357
|
3.641
|
|
Zfp868
|
zinc finger protein 868
|
|
chr13_-_114408704
|
3.624
|
NM_130796
|
Snx18
|
sorting nexin 18
|
|
chr6_+_124881376
|
3.605
|
NM_001170341
NM_145385
|
Mlf2
|
myeloid leukemia factor 2
|
|
chr15_+_6336747
|
3.581
|
NM_001037905
NM_001102400
NM_023118
|
Dab2
|
disabled homolog 2 (Drosophila)
|
|
chr5_-_67009992
|
3.564
|
|
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr2_+_167363696
|
3.546
|
NM_011427
|
Snai1
|
snail homolog 1 (Drosophila)
|
|
chr7_-_29941186
|
3.519
|
NM_001146023
|
Fam98c
|
family with sequence similarity 98, member C
|
|
chr11_-_118254729
|
3.499
|
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr10_+_25079661
|
3.499
|
|
Epb4.1l2
|
erythrocyte protein band 4.1-like 2
|
|
chr5_+_116009475
|
3.482
|
NM_007475
|
Rplp0
|
ribosomal protein, large, P0
|
|
chr11_+_83286635
|
3.472
|
|
Taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor
|
|
chr7_-_29678773
|
3.462
|
|
Actn4
|
actinin alpha 4
|
|
chr5_-_114578184
|
3.458
|
NM_175016
|
Alkbh2
|
alkB, alkylation repair homolog 2 (E. coli)
|
|
chr3_+_121948693
|
3.455
|
|
Gclm
|
glutamate-cysteine ligase, modifier subunit
|
|
chr14_+_66915914
|
3.437
|
|
Trim35
|
tripartite motif-containing 35
|
|
chr1_-_181448145
|
3.410
|
|
Smyd3
|
SET and MYND domain containing 3
|
|
chr19_-_38893670
|
3.406
|
NM_021315
|
Noc3l
|
nucleolar complex associated 3 homolog (S. cerevisiae)
|
|
chr8_+_73895190
|
3.386
|
NM_029865
|
Ocel1
|
occludin/ELL domain containing 1
|
|
chr4_+_151413427
|
3.373
|
NM_001159599
NM_028727
|
Nol9
|
nucleolar protein 9
|
|
chr10_-_14425271
|
3.366
|
NM_025418
|
Vta1
|
Vps20-associated 1 homolog (S. cerevisiae)
|