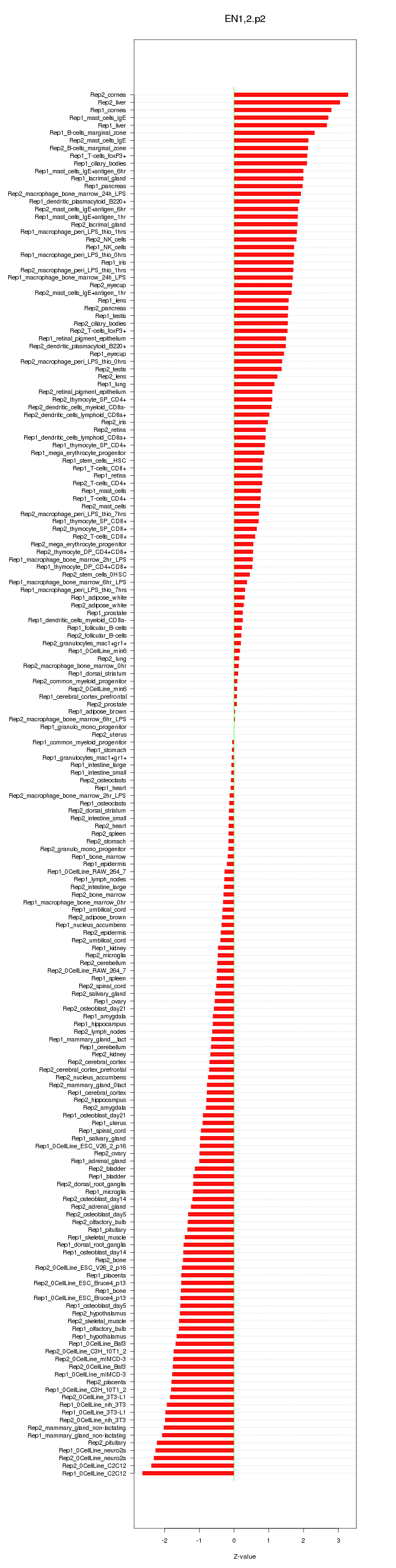
|
chr7_+_129776837
|
21.284
|
|
Prkcb
|
protein kinase C, beta
|
|
chr19_-_41406744
|
12.478
|
|
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr11_-_83462722
|
11.858
|
NM_011337
|
Ccl3
|
chemokine (C-C motif) ligand 3
|
|
chr6_-_87446228
|
11.243
|
NM_175476
|
Arhgap25
|
Rho GTPase activating protein 25
|
|
chr12_+_99506844
|
10.084
|
NM_008152
|
Gpr65
|
G-protein coupled receptor 65
|
|
chr7_-_27724396
|
9.436
|
NM_001101467
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22
|
|
chr2_-_126968325
|
8.872
|
NM_001163528
|
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr9_+_88851757
|
8.510
|
NM_009742
|
Bcl2a1a
|
B-cell leukemia/lymphoma 2 related protein A1a
|
|
chr6_+_83293435
|
8.466
|
|
Mobkl1b
|
MOB1, Mps One Binder kinase activator-like 1B (yeast)
|
|
chr7_+_52257572
|
8.369
|
|
|
|
|
chr7_+_52965052
|
8.340
|
|
Dbp
|
D site albumin promoter binding protein
|
|
chr1_+_174628164
|
8.160
|
NM_007768
|
Crp
|
C-reactive protein, pentraxin-related
|
|
chr2_-_129196855
|
7.982
|
NM_008361
|
Il1b
|
interleukin 1 beta
|
|
chr19_+_29446941
|
7.870
|
|
Cd274
|
CD274 antigen
|
|
chr19_-_34952328
|
7.744
|
|
Pank1
|
pantothenate kinase 1
|
|
chr18_-_77870871
|
7.697
|
|
2310007O11Rik
|
RIKEN cDNA 2310007O11 gene
|
|
chr8_+_41837561
|
7.472
|
NM_011781
|
Adam25
|
a disintegrin and metallopeptidase domain 25 (testase 2)
|
|
chr17_+_87762817
|
7.427
|
|
Ttc7
|
tetratricopeptide repeat domain 7
|
|
chr14_-_56143027
|
7.298
|
NM_008736
|
Nrl
|
neural retina leucine zipper gene
|
|
chr14_+_45607785
|
7.104
|
NM_008964
|
Ptger2
|
prostaglandin E receptor 2 (subtype EP2)
|
|
chr16_-_58928469
|
7.063
|
NM_146999
|
Olfr181
|
olfactory receptor 181
|
|
chr5_-_121262512
|
7.009
|
NM_033541
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C
|
|
chrX_+_104344792
|
6.934
|
NM_001122595
|
A630033H20Rik
|
RIKEN cDNA A630033H20 gene
|
|
chr14_-_56143799
|
6.923
|
NM_001136074
|
Nrl
|
neural retina leucine zipper gene
|
|
chr3_-_81779636
|
6.705
|
NM_019911
|
Tdo2
|
tryptophan 2,3-dioxygenase
|
|
chr19_-_34952377
|
6.605
|
NM_023792
|
Pank1
|
pantothenate kinase 1
|
|
chr8_+_72426082
|
6.517
|
NM_032004
|
Tssk6
|
testis-specific serine kinase 6
|
|
chr19_-_41068625
|
6.513
|
|
Blnk
|
B-cell linker
|
|
chr5_-_73143979
|
6.484
|
NM_001122754
|
Txk
|
TXK tyrosine kinase
|
|
chr7_-_53970378
|
6.483
|
|
Saa3
|
serum amyloid A 3
|
|
chr9_-_88626434
|
6.436
|
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr2_+_79547936
|
6.389
|
NM_172420
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr19_-_11698132
|
6.335
|
NM_013516
|
Ms4a2
|
membrane-spanning 4-domains, subfamily A, member 2
|
|
chr5_-_73127743
|
6.256
|
NM_013698
|
Txk
|
TXK tyrosine kinase
|
|
chr13_+_4190423
|
6.086
|
NM_013778
|
Akr1c13
|
aldo-keto reductase family 1, member C13
|
|
chr19_-_41069024
|
6.029
|
NM_008528
|
Blnk
|
B-cell linker
|
|
chr14_-_51766415
|
5.785
|
NM_011271
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr1_+_87918516
|
5.687
|
NM_001122732
NM_027029
NM_027300
NM_028647
|
Spata3
|
spermatogenesis associated 3
|
|
chr1_-_65109982
|
5.669
|
NM_007776
|
Crygd
|
crystallin, gamma D
|
|
chr11_-_99997241
|
5.650
|
NM_008469
|
Krt15
|
keratin 15
|
|
chrX_-_98463514
|
5.636
|
NM_013563
|
Il2rg
|
interleukin 2 receptor, gamma chain
|
|
chr11_-_99982808
|
5.551
|
NM_010662
|
Krt13
|
keratin 13
|
|
chr9_+_39201377
|
5.530
|
NM_001011812
|
Olfr951
|
olfactory receptor 951
|
|
chr2_+_121276592
|
5.477
|
|
Serf2
|
small EDRK-rich factor 2
|
|
chr4_+_32504409
|
5.463
|
NM_001109661
|
Bach2
|
BTB and CNC homology 2
|
|
chr11_-_77538789
|
5.440
|
NM_009965
|
Cryba1
|
crystallin, beta A1
|
|
chr10_-_53638995
|
5.423
|
|
Man1a
|
mannosidase 1, alpha
|
|
chr7_+_27620354
|
5.413
|
NM_007812
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5
|
|
chr10_+_128143313
|
5.403
|
NM_021882
|
Pmel
|
premelanosome protein
|
|
chr16_-_38800293
|
5.386
|
NM_178924
|
Upk1b
|
uroplakin 1B
|
|
chr1_-_80661844
|
5.268
|
|
Dock10
|
dedicator of cytokinesis 10
|
|
chrX_+_70962097
|
5.241
|
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr19_-_11340591
|
5.237
|
NM_007641
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chrX_+_104344265
|
5.166
|
NM_001122596
|
A630033H20Rik
|
RIKEN cDNA A630033H20 gene
|
|
chr11_+_49058980
|
5.148
|
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1
|
|
chr7_+_29222465
|
5.137
|
NM_022024
|
Gmfg
|
glia maturation factor, gamma
|
|
chr7_-_126666995
|
5.125
|
NM_027943
|
Pdilt
|
protein disulfide isomerase-like, testis expressed
|
|
chr6_+_39331379
|
5.075
|
NM_011226
|
Rab19
|
RAB19, member RAS oncogene family
|
|
chr8_+_107572489
|
4.936
|
NM_001164681
NM_198672
|
Ces3a
|
carboxylesterase 3A
|
|
chr5_-_86947601
|
4.892
|
NM_177162
|
9930032O22Rik
|
RIKEN cDNA 9930032O22 gene
|
|
chr8_-_107784507
|
4.819
|
|
Tradd
|
TNFRSF1A-associated via death domain
|
|
chr1_-_173537404
|
4.669
|
NM_008534
|
Ly9
|
lymphocyte antigen 9
|
|
chr5_-_104406333
|
4.656
|
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13
|
|
chr17_-_34134101
|
4.611
|
|
H2-K1
|
histocompatibility 2, K1, K region
|
|
chr17_-_34424623
|
4.525
|
NM_010378
|
H2-Aa
|
histocompatibility 2, class II antigen A, alpha
|
|
chr11_-_99236216
|
4.512
|
NM_027574
|
Krt28
|
keratin 28
|
|
chr7_+_27620380
|
4.510
|
|
Cyp2a4
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 4
cytochrome P450, family 2, subfamily a, polypeptide 5
|
|
chr16_+_90220986
|
4.506
|
NM_011434
|
Sod1
|
superoxide dismutase 1, soluble
|
|
chr6_-_115684968
|
4.492
|
NM_001168259
|
Tmem40
|
transmembrane protein 40
|
|
chr13_+_103483611
|
4.464
|
NM_008533
|
Cd180
|
CD180 antigen
|
|
chr6_+_89926164
|
4.458
|
NM_053229
|
Vmn1r46
|
vomeronasal 1 receptor 46
|
|
chr14_-_51815597
|
4.438
|
NM_007449
|
Ang2
|
angiogenin, ribonuclease A family, member 2
|
|
chr9_-_109751959
|
4.407
|
NM_009921
|
Camp
|
cathelicidin antimicrobial peptide
|
|
chr16_+_59148654
|
4.373
|
NM_146998
|
Olfr195
|
olfactory receptor 195
|
|
chr11_-_100957861
|
4.355
|
|
|
|
|
chr15_+_3230053
|
4.348
|
|
|
|
|
chr3_+_94736974
|
4.347
|
|
Selenbp1
|
selenium binding protein 1
|
|
chr2_+_14151018
|
4.335
|
NM_008625
|
Mrc1
|
mannose receptor, C type 1
|
|
chr11_-_95703752
|
4.298
|
NM_025659
NM_001163464
|
Abi3
|
ABI gene family, member 3
|
|
chr1_+_74421706
|
4.298
|
NM_013612
|
Slc11a1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1
|
|
chr16_+_17898729
|
4.290
|
NM_009436
|
Tssk2
|
testis-specific serine kinase 2
|
|
chr19_-_46747463
|
4.075
|
NM_007809
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1
|
|
chr17_-_85082214
|
3.969
|
NM_031884
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5
|
|
chr9_-_110959315
|
3.965
|
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr3_+_137949734
|
3.947
|
|
Adh1
|
alcohol dehydrogenase 1 (class I)
|
|
chr17_+_34052312
|
3.872
|
|
|
|
|
chr17_-_85082309
|
3.863
|
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5
|
|
chr17_-_34134309
|
3.801
|
|
H2-K1
|
histocompatibility 2, K1, K region
|
|
chr4_+_115845976
|
3.749
|
NM_001099295
|
1700042G07Rik
|
RIKEN cDNA 1700042G07 gene
|
|
chr8_-_44784840
|
3.730
|
NM_001079931
|
Adam34
Gm5347
|
a disintegrin and metallopeptidase domain 34
predicted gene 5347
|
|
chr9_-_88626613
|
3.708
|
NM_007536
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr16_+_20128178
|
3.684
|
|
Klhl24
|
kelch-like 24 (Drosophila)
|
|
chr11_-_99016390
|
3.657
|
NM_007719
|
Ccr7
|
chemokine (C-C motif) receptor 7
|
|
chr4_+_130469048
|
3.653
|
|
Laptm5
|
lysosomal-associated protein transmembrane 5
|
|
chr6_-_128776236
|
3.640
|
NM_030599
|
Klrb1b
|
killer cell lectin-like receptor subfamily B member 1B
|
|
chr7_+_27092187
|
3.632
|
NM_009997
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4
|
|
chr10_-_80131318
|
3.629
|
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2
|
|
chr1_+_154622471
|
3.628
|
|
|
|
|
chr17_+_13208071
|
3.525
|
|
Sod2
|
superoxide dismutase 2, mitochondrial
|
|
chr6_-_130287518
|
3.501
|
NM_010648
|
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3
|
|
chr10_-_111434231
|
3.487
|
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr16_-_30258448
|
3.472
|
|
|
|
|
chr5_-_92776027
|
3.468
|
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr6_-_129422702
|
3.460
|
NM_020008
|
Clec7a
|
C-type lectin domain family 7, member a
|
|
chr9_+_89094190
|
3.402
|
|
Bcl2a1a
|
B-cell leukemia/lymphoma 2 related protein A1a
|
|
chr2_-_17918493
|
3.388
|
NM_026627
|
H2afb1
|
H2A histone family, member B1
|
|
chr11_+_87895626
|
3.381
|
|
Vezf1
|
vascular endothelial zinc finger 1
|
|
chr16_-_48772068
|
3.357
|
NM_198297
|
Trat1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr17_+_12571494
|
3.348
|
|
Plg
|
plasminogen
|
|
chr14_+_56372605
|
3.312
|
|
2610027L16Rik
|
RIKEN cDNA 2610027L16 gene
|
|
chr12_+_34000269
|
3.249
|
|
Atxn7l1
|
ataxin 7-like 1
|
|
chr7_+_27814096
|
3.240
|
NM_133657
|
Cyp2a12
|
cytochrome P450, family 2, subfamily a, polypeptide 12
|
|
chr17_+_48594225
|
3.229
|
NM_152823
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like
|
|
chr5_+_88408469
|
3.220
|
NM_027631
|
4931407G18Rik
|
RIKEN cDNA 4931407G18 gene
|
|
chr5_-_115119973
|
3.216
|
|
Gltp
|
glycolipid transfer protein
|
|
chrX_+_70962057
|
3.161
|
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr3_+_138078450
|
3.143
|
NM_011996
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide
|
|
chr9_+_108411809
|
3.123
|
NM_001168270
|
Qars
|
glutaminyl-tRNA synthetase
|
|
chr10_+_127691272
|
3.067
|
|
Apon
|
apolipoprotein N
|
|
chr6_-_125186982
|
3.043
|
NM_001033126
NM_001042564
|
Cd27
|
CD27 antigen
|
|
chr9_+_4309898
|
3.036
|
|
Kbtbd3
|
kelch repeat and BTB (POZ) domain containing 3
|
|
chr16_+_20548675
|
3.020
|
NM_013852
|
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr15_+_4677207
|
3.019
|
NM_016704
|
C6
|
complement component 6
|
|
chr1_-_146024404
|
3.007
|
NM_153171
|
Rgs13
|
regulator of G-protein signaling 13
|
|
chr11_-_117630476
|
3.006
|
|
Tmc6
|
transmembrane channel-like gene family 6
|
|
chr5_-_121337533
|
3.005
|
NM_011852
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G
|
|
chr16_+_36935029
|
3.002
|
NM_008225
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1
|
|
chr11_-_99199267
|
2.966
|
NM_001033397
|
Krt26
|
keratin 26
|
|
chr19_+_10788964
|
2.950
|
|
|
|
|
chr14_-_47896531
|
2.947
|
NM_172598
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr6_-_119305193
|
2.944
|
|
Adipor2
|
adiponectin receptor 2
|
|
chr3_-_123375305
|
2.938
|
NM_031186
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr9_-_110959702
|
2.902
|
NM_017466
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr2_+_92223074
|
2.886
|
NM_183112
|
1700029I15Rik
|
RIKEN cDNA 1700029I15 gene
|
|
chr2_-_126969008
|
2.870
|
NM_001163527
|
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr19_-_29095519
|
2.856
|
|
|
|
|
chr3_-_86803173
|
2.855
|
NM_007639
|
Cd1d1
|
CD1d1 antigen
|
|
chr17_+_25503264
|
2.842
|
NM_010781
|
Tpsb2
|
tryptase beta 2
|
|
chr14_+_21513864
|
2.805
|
|
|
|
|
chr7_-_51787440
|
2.801
|
NM_019866
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr1_+_173144142
|
2.800
|
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3
|
|
chr10_+_111602731
|
2.791
|
NM_178278
|
Caps2
|
calcyphosphine 2
|
|
chr13_-_93346126
|
2.789
|
NM_030237
|
Spz1
|
spermatogenic leucine zipper 1
|
|
chr11_+_95703598
|
2.772
|
NM_023121
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr4_+_147976641
|
2.765
|
NM_001003893
NM_010767
|
Masp2
|
mannan-binding lectin serine peptidase 2
|
|
chr5_-_124315478
|
2.761
|
NM_030701
|
Niacr1
|
niacin receptor 1
|
|
chrX_-_5897723
|
2.740
|
NM_009757
|
Bmp15
|
bone morphogenetic protein 15
|
|
chr10_-_24647173
|
2.725
|
|
Arg1
|
arginase, liver
|
|
chr5_-_92682184
|
2.688
|
NM_011148
|
Ppef2
|
protein phosphatase, EF hand calcium-binding domain 2
|
|
chr13_-_21384553
|
2.685
|
NM_010343
|
Gpx5
|
glutathione peroxidase 5
|
|
chr17_+_12571455
|
2.678
|
NM_008877
|
Plg
|
plasminogen
|
|
chr3_+_54943959
|
2.638
|
NM_177203
|
A730037C10Rik
|
RIKEN cDNA A730037C10 gene
|
|
chr3_-_58984006
|
2.635
|
NM_032399
|
Gpr87
|
G protein-coupled receptor 87
|
|
chr2_-_26314921
|
2.600
|
|
Notch1
|
Notch gene homolog 1 (Drosophila)
|
|
chr6_+_15750966
|
2.593
|
|
Mdfic
|
MyoD family inhibitor domain containing
|
|
chr11_+_105836521
|
2.589
|
NM_009598
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr8_-_44613490
|
2.579
|
NM_001009547
|
Adam26b
|
a disintegrin and metallopeptidase domain 26B
|
|
chr14_-_31756446
|
2.576
|
NM_008406
|
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1
|
|
chr13_-_4149873
|
2.562
|
NM_134066
|
Akr1c18
|
aldo-keto reductase family 1, member C18
|
|
chr15_+_84155149
|
2.558
|
NM_022321
|
Parvg
|
parvin, gamma
|
|
chr15_+_78156227
|
2.528
|
NM_007780
|
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr4_+_48064119
|
2.509
|
NM_015743
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr9_-_44611033
|
2.494
|
NM_138951
|
Ttc36
|
tetratricopeptide repeat domain 36
|
|
chr7_-_5148348
|
2.483
|
NM_030740
|
Vmn1r56
|
vomeronasal 1 receptor 56
|
|
chr6_+_57531124
|
2.478
|
|
Herc6
|
hect domain and RLD 6
|
|
chr11_-_72609306
|
2.469
|
NM_001024926
|
Cyb5d2
|
cytochrome b5 domain containing 2
|
|
chr2_+_25312352
|
2.469
|
|
Clic3
|
chloride intracellular channel 3
|
|
chr11_-_69619087
|
2.464
|
NM_001033433
|
Tmem102
|
transmembrane protein 102
|
|
chr2_-_103105045
|
2.458
|
|
Ehf
|
ets homologous factor
|
|
chr14_-_47896464
|
2.447
|
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr5_-_66472208
|
2.436
|
|
Rbm47
|
RNA binding motif protein 47
|
|
chr10_-_117732093
|
2.410
|
NM_054079
|
Iltifb
|
interleukin 10-related T cell-derived inducible factor beta
|
|
chr16_-_58872973
|
2.406
|
NM_146993
NM_146996
|
Olfr176
Olfr177
|
olfactory receptor 176
olfactory receptor 177
|
|
chr7_-_147174332
|
2.369
|
|
Adam8
|
a disintegrin and metallopeptidase domain 8
|
|
chr7_-_127813763
|
2.363
|
NM_001040137
NM_001040136
|
Pdzd9
|
PDZ domain containing 9
|
|
chr9_-_123883398
|
2.345
|
NM_009912
|
Ccr1
|
chemokine (C-C motif) receptor 1
|
|
chr1_-_175157322
|
2.339
|
NM_010184
|
Fcer1a
|
Fc receptor, IgE, high affinity I, alpha polypeptide
|
|
chr14_+_56590264
|
2.326
|
NM_001024714
|
Cma2
|
chymase 2, mast cell
|
|
chr14_+_56188462
|
2.317
|
|
|
|
|
chr3_-_135988955
|
2.309
|
NM_001033350
|
Bank1
|
B-cell scaffold protein with ankyrin repeats 1
|
|
chr9_-_80359017
|
2.304
|
NM_022016
|
Impg1
|
interphotoreceptor matrix proteoglycan 1
|
|
chr10_+_127691166
|
2.277
|
NM_133996
|
Apon
|
apolipoprotein N
|
|
chr4_+_147415297
|
2.261
|
NM_001161798
|
Mthfr
|
5,10-methylenetetrahydrofolate reductase
|
|
chr14_+_32481114
|
2.260
|
|
Btd
|
biotinidase
|
|
chr4_-_106472351
|
2.255
|
NM_025590
|
Acot11
|
acyl-CoA thioesterase 11
|
|
chr6_-_129335520
|
2.254
|
NM_027709
|
Clec12b
|
C-type lectin domain family 12, member B
|
|
chr2_+_25312358
|
2.238
|
NM_027085
|
Clic3
|
chloride intracellular channel 3
|
|
chr7_-_111042984
|
2.233
|
NM_013617
|
Olfr64
|
olfactory receptor 64
|
|
chr4_-_19849712
|
2.231
|
NM_175406
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2
|
|
chr3_+_59729135
|
2.229
|
NM_177661
|
C130079G13Rik
|
RIKEN cDNA C130079G13 gene
|
|
chr6_+_41488583
|
2.207
|
|
|
|
|
chr17_+_35610430
|
2.205
|
|
H2-Q10
|
histocompatibility 2, Q region locus 10
|
|
chr14_-_64162562
|
2.204
|
|
Mtmr9
|
myotubularin related protein 9
|
|
chr2_+_30122256
|
2.197
|
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr3_-_20174589
|
2.183
|
NM_029706
|
Cpb1
|
carboxypeptidase B1 (tissue)
|
|
chr17_+_34335139
|
2.171
|
NM_010724
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7)
|
|
chr6_+_142246944
|
2.166
|
NM_010491
|
Iapp
|
islet amyloid polypeptide
|
|
chr3_+_90472990
|
2.165
|
NM_013650
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A)
|
|
chr16_+_45224449
|
2.154
|
NM_001037719
NM_177584
|
Btla
|
B and T lymphocyte associated
|
|
chr14_+_47896669
|
2.154
|
NM_080843
|
Socs4
|
suppressor of cytokine signaling 4
|