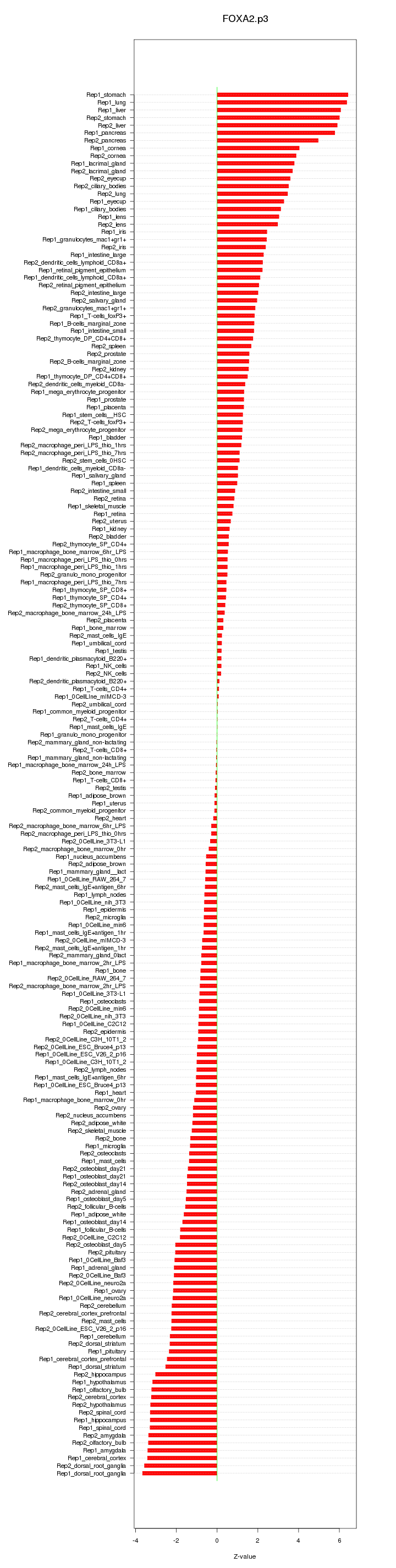
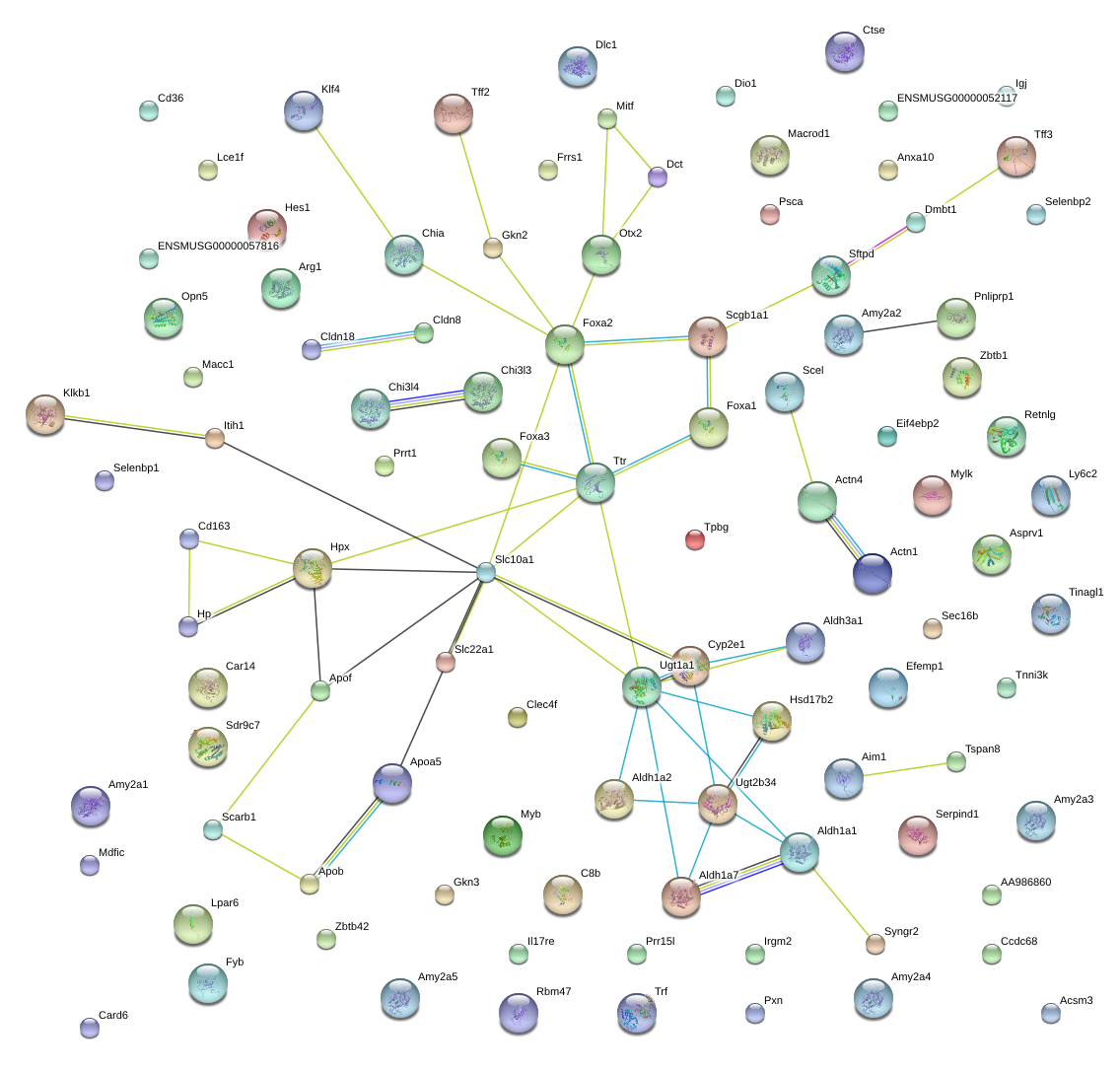
|
chr3_-_113061599
|
39.206
|
NM_001042711
|
Amy2a5
|
amylase 2a5
|
|
chr3_-_113094211
|
35.120
|
NM_001160150
NM_001160151
NM_001160152
NM_001042711
|
Amy2a4
Amy2a3
Amy2a2
Amy2a5
|
amylase 2a4
amylase 2a3
amylase 2a2
amylase 2a5
|
|
chr3_-_113126814
|
30.738
|
NM_001160150
NM_001160151
NM_001160152
NM_001042711
|
Amy2a4
Amy2a3
Amy2a2
Amy2a5
|
amylase 2a4
amylase 2a3
amylase 2a2
amylase 2a5
|
|
chr7_+_138175581
|
30.059
|
NM_007769
|
Dmbt1
|
deleted in malignant brain tumors 1
|
|
chr14_-_118450875
|
26.965
|
NM_010024
|
Dct
|
dopachrome tautomerase
|
|
chr12_-_58646914
|
22.464
|
|
Foxa1
|
forkhead box A1
|
|
chr4_+_104438905
|
20.993
|
NM_133882
|
C8b
|
complement component 8, beta polypeptide
|
|
chr10_-_24647243
|
20.883
|
NM_007482
|
Arg1
|
arginase, liver
|
|
chr3_-_113159420
|
18.704
|
NM_001160150
NM_001160151
NM_001160152
NM_001042711
|
Amy2a4
Amy2a3
Amy2a2
Amy2a5
|
amylase 2a4
amylase 2a3
amylase 2a2
amylase 2a5
|
|
chr15_+_74545268
|
17.991
|
NM_028216
|
Psca
|
prostate stem cell antigen
|
|
chr19_-_9162377
|
17.527
|
NM_011681
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr16_-_88563421
|
17.509
|
NM_018778
|
Cldn8
|
claudin 8
|
|
chr14_+_103912557
|
17.310
|
NM_022886
|
Scel
|
sciellin
|
|
chr8_+_120225803
|
17.205
|
NM_008290
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr5_-_87335956
|
17.190
|
NM_153598
|
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34
|
|
chr6_+_87323350
|
16.793
|
NM_025467
|
Gkn2
|
gastrokine 2
|
|
chr17_-_31281184
|
16.416
|
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1)
|
|
chr4_-_55545337
|
16.328
|
NM_010637
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr6_+_86578550
|
16.190
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr11_+_96790581
|
16.137
|
NM_146026
|
Prr15l
|
proline rich 15-like
|
|
chr16_-_88563379
|
15.990
|
|
Cldn8
|
claudin 8
|
|
chr4_-_55545211
|
15.807
|
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr17_-_31281223
|
15.551
|
NM_009363
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1)
|
|
chr11_+_28821585
|
15.448
|
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1
|
|
chr5_-_88956788
|
15.287
|
NM_152839
|
Igj
|
immunoglobulin joining chain
|
|
chr4_-_57929087
|
14.956
|
NM_178727
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene
|
|
chr3_-_112959644
|
14.623
|
NM_001190403
NM_001190404
|
Amy2b
|
amylase 2b
|
|
chr6_+_86578269
|
14.486
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr15_-_5058532
|
14.276
|
NM_001163138
|
Card6
|
caspase recruitment domain family, member 6
|
|
chr5_-_66472208
|
14.182
|
|
Rbm47
|
RNA binding motif protein 47
|
|
chr18_+_20823908
|
13.733
|
|
Ttr
|
transthyretin
|
|
chr8_-_46380114
|
13.704
|
|
Klkb1
|
kallikrein B, plasma 1
|
|
chr8_-_46380165
|
13.591
|
NM_008455
|
Klkb1
|
kallikrein B, plasma 1
|
|
chr1_+_89999782
|
13.566
|
NM_201410
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B
|
|
chr6_+_70676428
|
13.357
|
|
|
|
|
chr9_-_99617685
|
13.335
|
NM_001194921
NM_001194923
|
Cldn18
|
claudin 18
|
|
chr8_+_120225866
|
13.267
|
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr1_+_159456274
|
13.047
|
NM_001159986
|
Sec16b
|
SEC16 homolog B (S. cerevisiae)
|
|
chr9_-_99610427
|
12.921
|
|
Cldn18
|
claudin 18
|
|
chr17_-_12868558
|
12.858
|
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1
|
|
chr18_+_70085202
|
12.720
|
NM_201362
|
Ccdc68
|
coiled-coil domain containing 68
|
|
chrX_-_35772763
|
12.639
|
|
|
|
|
chr12_+_113917038
|
12.607
|
NM_001100460
|
Zbtb42
|
zinc finger and BTB domain containing 42
|
|
chr17_-_12868607
|
12.508
|
NM_009202
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1
|
|
chr9_-_99610481
|
11.963
|
NM_001194922
NM_019815
|
Cldn18
|
claudin 18
|
|
chr10_-_20880559
|
11.891
|
NM_001198914
NM_010848
|
Myb
|
myeloblastosis oncogene
|
|
chr12_-_82068951
|
11.592
|
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1
|
|
chr6_+_86578225
|
11.579
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr4_-_57928618
|
11.453
|
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene
|
|
chr14_-_49282546
|
11.348
|
NM_144841
|
Otx2
|
orthodenticle homolog 2 (Drosophila)
|
|
chr1_+_133558507
|
11.327
|
|
Ctse
|
cathepsin E
|
|
chr16_+_35002066
|
11.300
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr1_+_133558486
|
11.243
|
|
Ctse
|
cathepsin E
|
|
chr5_-_17394564
|
11.203
|
|
Cd36
|
CD36 antigen
|
|
chr8_-_112102946
|
10.956
|
|
Hp
|
haptoglobin
|
|
chr3_+_116564564
|
10.863
|
NM_009146
|
Frrs1
|
ferric-chelate reductase 1
|
|
chr6_+_113408477
|
10.847
|
NM_145826
|
Il17re
|
interleukin 17 receptor E
|
|
chr16_+_48872720
|
10.818
|
NM_181596
|
Retnlg
|
resistin like gamma
|
|
chr8_-_64601861
|
10.725
|
NM_001136089
NM_011922
|
Anxa10
|
annexin A10
|
|
chr3_-_154718282
|
10.686
|
NM_177066
|
Tnni3k
|
TNNI3 interacting kinase
|
|
chr10_+_115254308
|
10.484
|
NM_001168679
NM_001168680
NM_146010
|
Tspan8
|
tetraspanin 8
|
|
chr6_+_70676803
|
10.473
|
|
|
|
|
chr19_+_7131270
|
10.470
|
|
Macrod1
|
MACRO domain containing 1
|
|
chr12_-_82069060
|
10.325
|
NM_001177561
NM_011387
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1
|
|
chr14_+_73637961
|
10.305
|
|
Lpar6
|
lysophosphatidic acid receptor 6
|
|
chr1_+_133558517
|
10.288
|
|
Ctse
|
cathepsin E
|
|
chr6_+_97941776
|
10.228
|
NM_008601
|
Mitf
|
microphthalmia-associated transcription factor
|
|
chr3_+_105916283
|
10.225
|
NM_023186
|
Chia
|
chitinase, acidic
|
|
chr11_+_117675276
|
10.056
|
|
Syngr2
|
synaptogyrin 2
|
|
chr6_+_86578167
|
9.995
|
NM_026414
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr8_-_112102961
|
9.963
|
|
Hp
|
haptoglobin
|
|
chr19_+_7131325
|
9.953
|
|
Macrod1
|
MACRO domain containing 1
|
|
chr6_-_83606025
|
9.929
|
|
Clec4f
|
C-type lectin domain family 4, member f
|
|
chr18_+_20823750
|
9.872
|
NM_013697
|
Ttr
|
transthyretin
|
|
chr14_-_31756446
|
9.806
|
NM_008406
|
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1
|
|
chr12_+_7984451
|
9.793
|
NM_009693
|
Apob
|
apolipoprotein B
|
|
chr1_+_90097175
|
9.733
|
NM_013701
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2
|
|
chr7_-_112748585
|
9.708
|
|
Hpx
|
hemopexin
|
|
chr1_+_132628552
|
9.598
|
NM_177604
|
AA986860
|
expressed sequence AA986860
|
|
chr6_-_83606100
|
9.559
|
NM_016751
|
Clec4f
|
C-type lectin domain family 4, member f
|
|
chr7_-_112748624
|
9.507
|
NM_017371
|
Hpx
|
hemopexin
|
|
chr4_-_106979732
|
9.497
|
NM_007860
|
Dio1
|
deiodinase, iodothyronine, type I
|
|
chr14_+_73637697
|
9.490
|
NM_175116
|
Lpar6
|
lysophosphatidic acid receptor 6
|
|
chr16_+_30065419
|
9.463
|
NM_008235
|
Hes1
|
hairy and enhancer of split 1 (Drosophila)
|
|
chr12_+_120681882
|
9.446
|
NM_001163136
|
Macc1
|
metastasis associated in colon cancer 1
|
|
chr3_+_94736993
|
9.432
|
NM_009150
|
Selenbp1
|
selenium binding protein 1
|
|
chr14_-_41998434
|
9.396
|
NM_009160
|
Sftpd
|
surfactant associated protein D
|
|
chr7_+_126904427
|
9.365
|
NM_016870
NM_212441
NM_212442
|
Acsm3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr6_+_15750966
|
9.340
|
|
Mdfic
|
MyoD family inhibitor domain containing
|
|
chr9_+_46076832
|
9.290
|
|
Apoa5
|
apolipoprotein A-V
|
|
chr6_-_87338847
|
9.281
|
NM_026860
|
Gkn3
|
gastrokine 3
|
|
chr3_+_94736974
|
9.141
|
|
Selenbp1
|
selenium binding protein 1
|
|
chr11_+_58028487
|
9.132
|
|
Irgm2
|
immunity-related GTPase family M member 2
|
|
chr11_+_61022240
|
9.052
|
NM_001112725
NM_007436
|
Aldh3a1
|
aldehyde dehydrogenase family 3, subfamily A1
|
|
chr19_+_58803376
|
9.027
|
NM_018874
|
Pnliprp1
|
pancreatic lipase related protein 1
|
|
chr7_-_29678773
|
8.960
|
|
Actn4
|
actinin alpha 4
|
|
chr3_+_94497485
|
8.905
|
NM_019414
|
Selenbp2
|
selenium binding protein 2
|
|
chr5_+_116005777
|
8.887
|
|
Pxn
|
paxillin
|
|
chr14_+_79601537
|
8.878
|
|
|
|
|
chr15_+_6529846
|
8.722
|
NM_011815
|
Fyb
|
FYN binding protein
|
|
chr12_+_77471506
|
8.613
|
|
Zbtb1
|
zinc finger and BTB domain containing 1
|
|
chr10_+_127705664
|
8.600
|
|
Apof
|
apolipoprotein F
|
|
chr16_+_17331463
|
8.593
|
NM_008223
|
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1
|
|
chr10_+_127335573
|
8.556
|
NM_027301
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7
|
|
chr3_-_95708543
|
8.460
|
NM_011797
|
Car14
|
carbonic anhydrase 14
|
|
chr16_+_17331507
|
8.375
|
|
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1
|
|
chr5_-_17355533
|
8.352
|
NM_001159556
NM_001159557
|
Cd36
|
CD36 antigen
|
|
chr7_+_147949637
|
8.265
|
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1
|
|
chr19_-_20802015
|
8.237
|
NM_011921
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7
|
|
chr13_-_38069696
|
8.195
|
|
|
|
|
chr17_+_34766489
|
8.194
|
NM_030890
|
Prrt1
|
proline-rich transmembrane protein 1
|
|
chr10_-_60915251
|
8.181
|
NM_010124
|
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr1_-_135972341
|
8.155
|
|
|
|
|
chr3_-_105970438
|
8.008
|
NM_009892
|
Chi3l3
|
chitinase 3-like 3
|
|
chr6_+_124254668
|
7.906
|
NM_001170395
NM_053094
|
Cd163
|
CD163 antigen
|
|
chr6_+_90424727
|
7.896
|
|
Klf15
|
Kruppel-like factor 15
|
|
chr6_-_87793459
|
7.860
|
|
Cnbp
|
cellular nucleic acid binding protein
|
|
chr19_+_55816299
|
7.834
|
NM_001142918
NM_001142919
NM_001142920
NM_001142921
NM_001142922
NM_001142923
NM_009333
|
Tcf7l2
|
transcription factor 7-like 2, T-cell specific, HMG-box
|
|
chr11_+_110260435
|
7.793
|
NM_011943
|
Map2k6
|
mitogen-activated protein kinase kinase 6
|
|
chr3_+_106288967
|
7.790
|
NM_028110
|
Dennd2d
|
DENN/MADD domain containing 2D
|
|
chr6_-_3444484
|
7.778
|
NM_178899
|
Hepacam2
|
HEPACAM family member 2
|
|
chr10_+_23571267
|
7.773
|
NM_011979
|
Vnn3
|
vanin 3
|
|
chr9_+_53109774
|
7.771
|
NM_176846
|
Exph5
|
exophilin 5
|
|
chr6_-_5143755
|
7.766
|
NM_011134
|
Pon1
|
paraoxonase 1
|
|
chr3_-_115710131
|
7.746
|
NM_023214
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr7_-_94641859
|
7.558
|
NM_011661
|
Tyr
|
tyrosinase
|
|
chrX_+_157205944
|
7.558
|
NM_011302
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human)
|
|
chr1_+_45402907
|
7.531
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr12_+_77471173
|
7.528
|
NM_178744
|
Zbtb1
|
zinc finger and BTB domain containing 1
|
|
chr6_+_135312948
|
7.413
|
NM_010128
|
Emp1
|
epithelial membrane protein 1
|
|
chr9_+_46076689
|
7.396
|
NM_080434
|
Apoa5
|
apolipoprotein A-V
|
|
chr15_-_102008126
|
7.388
|
|
|
|
|
chr6_+_87040212
|
7.344
|
|
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1
|
|
chr16_+_22892112
|
7.338
|
NM_013465
|
Ahsg
|
alpha-2-HS-glycoprotein
|
|
chr6_-_134741351
|
7.329
|
|
Dusp16
|
dual specificity phosphatase 16
|
|
chr1_+_89951985
|
7.284
|
NM_201641
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10
|
|
chr11_+_94426358
|
7.255
|
NM_007689
|
Chad
|
chondroadherin
|
|
chr3_-_131047840
|
7.249
|
NM_028943
|
Sgms2
|
sphingomyelin synthase 2
|
|
chr7_+_109378512
|
7.234
|
|
Pgap2
|
post-GPI attachment to proteins 2
|
|
chr17_+_34400152
|
7.212
|
NM_207105
|
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1
|
|
chr10_+_127142225
|
7.161
|
NM_001081219
|
Myo1a
|
myosin IA
|
|
chr5_+_65221116
|
7.160
|
|
Klf3
|
Kruppel-like factor 3 (basic)
|
|
chr8_-_112103065
|
7.091
|
NM_017370
|
Hp
|
haptoglobin
|
|
chr6_+_135313010
|
7.050
|
|
Emp1
|
epithelial membrane protein 1
|
|
chr16_+_22892125
|
6.977
|
|
Ahsg
|
alpha-2-HS-glycoprotein
|
|
chr15_+_54242324
|
6.920
|
NM_173422
|
Colec10
|
collectin sub-family member 10
|
|
chr3_-_20174589
|
6.912
|
NM_029706
|
Cpb1
|
carboxypeptidase B1 (tissue)
|
|
chrX_-_93488752
|
6.859
|
NM_177789
|
Vsig4
|
V-set and immunoglobulin domain containing 4
|
|
chr19_-_44103644
|
6.810
|
NM_001001446
NM_001167905
|
Cyp2c44
|
cytochrome P450, family 2, subfamily c, polypeptide 44
|
|
chr1_-_138856866
|
6.803
|
NM_030676
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr10_-_127325668
|
6.800
|
|
Rdh7
|
retinol dehydrogenase 7
|
|
chr10_-_88996314
|
6.779
|
NM_001163504
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr11_+_87895626
|
6.735
|
|
Vezf1
|
vascular endothelial zinc finger 1
|
|
chr7_+_52976589
|
6.726
|
NM_001033170
|
Fam83e
|
family with sequence similarity 83, member E
|
|
chr3_-_144695718
|
6.677
|
NM_017474
|
Clca3
|
chloride channel calcium activated 3
|
|
chr19_+_7131234
|
6.648
|
NM_134147
|
Macrod1
|
MACRO domain containing 1
|
|
chr16_+_33518414
|
6.618
|
NM_134251
|
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr17_-_7997838
|
6.598
|
NM_001081416
|
Fndc1
|
fibronectin type III domain containing 1
|
|
chr10_+_127691272
|
6.588
|
|
Apon
|
apolipoprotein N
|
|
chr3_+_19857053
|
6.588
|
NM_001042611
NM_007752
|
Cp
|
ceruloplasmin
|
|
chr5_-_91069499
|
6.570
|
NM_028478
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr16_-_22811458
|
6.559
|
NM_009967
|
Crygs
|
crystallin, gamma S
|
|
chr10_-_24647173
|
6.553
|
|
Arg1
|
arginase, liver
|
|
chrX_+_160578411
|
6.479
|
NM_027286
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr3_+_96636596
|
6.439
|
NM_001146001
|
Pdzk1
|
PDZ domain containing 1
|
|
chr16_+_37580238
|
6.434
|
NM_013547
|
Hgd
|
homogentisate 1, 2-dioxygenase
|
|
chr16_+_33500899
|
6.418
|
|
Zfp148
|
zinc finger protein 148
|
|
chr2_+_153834000
|
6.416
|
NM_008953
|
Psp
|
parotid secretory protein
|
|
chr2_-_27118325
|
6.413
|
|
Vav2
|
vav 2 oncogene
|
|
chrX_-_35793335
|
6.383
|
|
Lamp2
|
lysosomal-associated membrane protein 2
|
|
chr19_-_38117813
|
6.380
|
|
Myof
|
myoferlin
|
|
chr10_-_117252126
|
6.376
|
|
|
|
|
chr2_-_60560659
|
6.370
|
NM_001159564
|
Itgb6
|
integrin beta 6
|
|
chr2_-_62321694
|
6.331
|
NM_008100
|
Gcg
|
glucagon
|
|
chr16_+_22892156
|
6.314
|
|
Ahsg
|
alpha-2-HS-glycoprotein
|
|
chr3_+_135488454
|
6.307
|
NM_026228
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8
|
|
chr5_-_146691425
|
6.296
|
NM_007818
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11
|
|
chr7_+_120909680
|
6.256
|
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein
|
|
chr17_+_12571494
|
6.235
|
|
Plg
|
plasminogen
|
|
chr3_-_8964045
|
6.232
|
NM_001025261
NM_001025262
|
Tpd52
|
tumor protein D52
|
|
chr4_-_49562319
|
6.227
|
NM_144903
|
Aldob
|
aldolase B, fructose-bisphosphate
|
|
chr6_+_34659440
|
6.225
|
NM_145575
|
Cald1
|
caldesmon 1
|
|
chr10_+_42580317
|
6.191
|
NM_172938
|
Scml4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr7_-_77511242
|
6.173
|
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_-_81779636
|
6.170
|
NM_019911
|
Tdo2
|
tryptophan 2,3-dioxygenase
|
|
chr11_-_31724523
|
6.161
|
|
D630024D03Rik
|
RIKEN cDNA D630024D03 gene
|
|
chr16_+_34995000
|
6.151
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr19_+_20676453
|
6.104
|
NM_013467
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1
|
|
chr13_+_4190423
|
6.093
|
NM_013778
|
Akr1c13
|
aldo-keto reductase family 1, member C13
|
|
chr10_-_78180902
|
6.083
|
NM_009809
|
Casp14
|
caspase 14
|
|
chr6_+_30491641
|
6.055
|
NM_001024698
|
Cpa2
|
carboxypeptidase A2, pancreatic
|
|
chr1_+_140864297
|
6.051
|
NM_181347
|
Dennd1b
|
DENN/MADD domain containing 1B
|
|
chr1_-_138130840
|
6.044
|
NM_028872
|
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene
|
|
chr9_-_44611055
|
6.038
|
|
Ttc36
|
tetratricopeptide repeat domain 36
|
|
chr3_+_138078450
|
6.024
|
NM_011996
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide
|
|
chr1_+_45390415
|
6.022
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr19_-_34329804
|
6.009
|
NM_007392
|
Acta2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr17_+_12571455
|
5.929
|
NM_008877
|
Plg
|
plasminogen
|
|
chr12_-_104976324
|
5.925
|
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B
|
|
chr11_+_3931731
|
5.798
|
NM_146013
|
Sec14l4
|
SEC14-like 4 (S. cerevisiae)
|