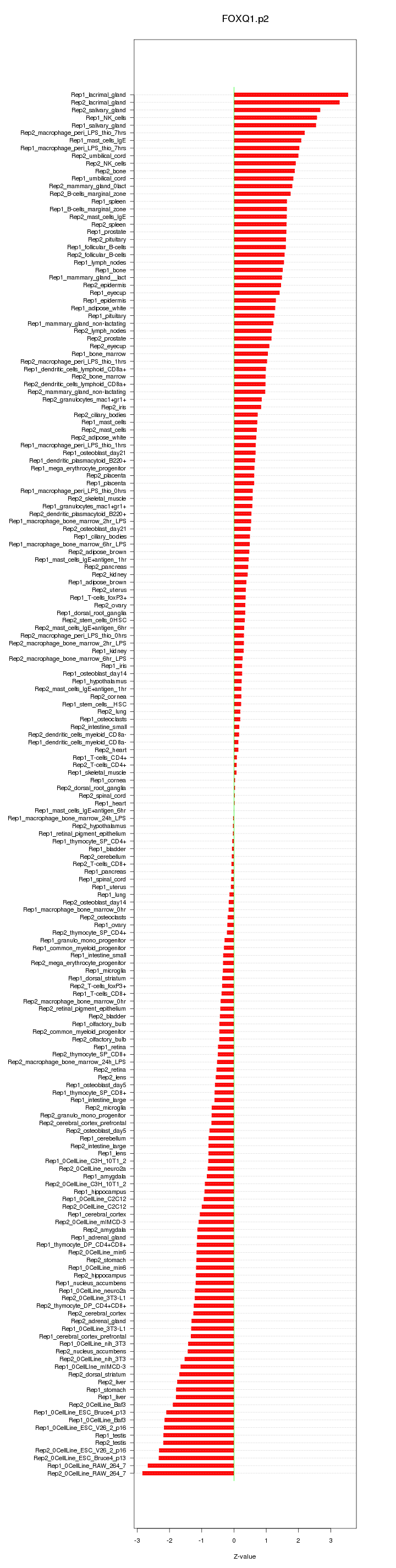
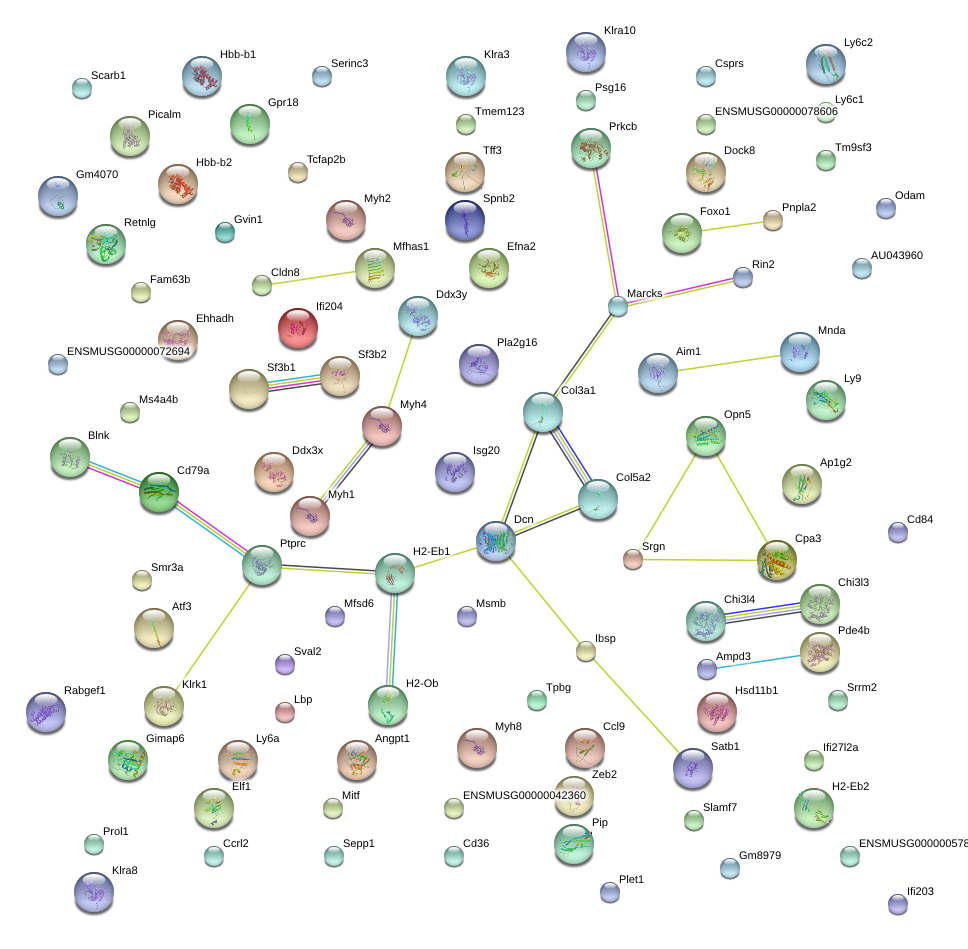
|
chr9_+_7792286
|
15.452
|
|
Tmem123
|
transmembrane protein 123
|
|
chr8_-_124948837
|
14.589
|
|
|
|
|
chr1_-_140071818
|
14.492
|
NM_001111316
NM_011210
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_-_100678555
|
14.352
|
|
Xist
|
inactive X specific transcripts
|
|
chr17_-_51878720
|
14.057
|
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_+_34442808
|
12.726
|
NM_010382
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr17_+_34442837
|
12.348
|
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr11_-_83388330
|
11.831
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr10_+_96942124
|
10.514
|
NM_001190451
|
Dcn
|
decorin
|
|
chr9_+_50302624
|
9.620
|
NM_029639
|
1600029D21Rik
|
RIKEN cDNA 1600029D21 gene
|
|
chr16_+_43321404
|
9.580
|
|
|
|
|
chr14_-_122314994
|
8.584
|
NM_182806
|
Gpr18
|
G protein-coupled receptor 18
|
|
chr6_-_48658182
|
7.976
|
|
Gimap6
|
GTPase, IMAP family member 6
|
|
chr1_+_173770709
|
7.956
|
NM_013489
|
Cd84
|
CD84 antigen
|
|
chr10_+_96944961
|
7.947
|
NM_007833
|
Dcn
|
decorin
|
|
chr1_-_175805812
|
7.867
|
|
Mndal
|
myeloid nuclear differentiation antigen like
|
|
chr14_+_79915429
|
7.522
|
|
Elf1
|
E74-like factor 1
|
|
chr4_+_102243132
|
7.508
|
|
Pde4b
|
phosphodiesterase 4B, cAMP specific
|
|
chr5_+_88746336
|
7.275
|
NM_008644
|
Prol1
|
proline rich, lacrimal 1
|
|
chr7_+_148645664
|
7.133
|
|
Pnpla2
|
patatin-like phospholipase domain containing 2
|
|
chr1_-_140071630
|
7.045
|
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C
|
|
chr5_-_17341534
|
6.987
|
|
Cd36
|
CD36 antigen
|
|
chr1_-_195068455
|
6.939
|
NM_001044751
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1
|
|
chr6_-_48658211
|
6.880
|
NM_153175
|
Gimap6
|
GTPase, IMAP family member 6
|
|
chr15_-_74828317
|
6.813
|
NM_010738
|
Ly6a
|
lymphocyte antigen 6 complex, locus A
|
|
chr6_+_97941776
|
6.761
|
NM_008601
|
Mitf
|
microphthalmia-associated transcription factor
|
|
chr3_-_105970438
|
6.646
|
NM_009892
|
Chi3l3
|
chitinase 3-like 3
|
|
chr7_+_117916117
|
6.634
|
NM_009667
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr7_+_129776837
|
6.628
|
|
Prkcb
|
protein kinase C, beta
|
|
chr15_-_74879242
|
6.613
|
NM_010741
|
Ly6c1
|
lymphocyte antigen 6 complex, locus C1
|
|
chr7_+_97357406
|
6.591
|
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_-_113358823
|
6.572
|
NM_001039160
NM_029000
|
Gvin1
|
GTPase, very large interferon inducible 1
|
|
chr7_-_113102453
|
6.572
|
NM_001039160
NM_029000
|
Gvin1
|
GTPase, very large interferon inducible 1
|
|
chr1_+_45390415
|
6.511
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr16_-_88563421
|
6.457
|
NM_018778
|
Cldn8
|
claudin 8
|
|
chr7_+_86058650
|
6.393
|
NM_020583
|
Isg20
|
interferon-stimulated protein
|
|
chrX_+_12870471
|
6.389
|
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked
|
|
chr1_-_192994532
|
6.386
|
|
Atf3
|
activating transcription factor 3
|
|
chr7_+_86058817
|
6.194
|
|
Isg20
|
interferon-stimulated protein
|
|
chr7_+_17659388
|
6.134
|
NM_007676
|
Psg16
|
pregnancy specific glycoprotein 16
|
|
chr10_-_61960613
|
6.130
|
|
Srgn
|
serglycin
|
|
chr17_+_23949101
|
6.102
|
|
Srrm2
|
serine/arginine repetitive matrix 2
|
|
chr6_+_41810237
|
6.081
|
NM_032542
|
Sval2
|
seminal vesicle antigen-like 2
|
|
chr15_+_3230053
|
6.068
|
|
|
|
|
chr1_-_173579128
|
6.054
|
|
Slamf7
|
SLAM family member 7
|
|
chr16_-_88563379
|
6.015
|
|
Cldn8
|
claudin 8
|
|
chr2_+_13504271
|
6.003
|
|
|
|
|
chr10_-_36855292
|
5.938
|
|
Marcks
|
myristoylated alanine rich protein kinase C substrate
|
|
chr17_+_34375847
|
5.902
|
NM_010389
|
H2-Ob
|
histocompatibility 2, O region beta locus
|
|
chr3_+_52153283
|
5.788
|
|
Foxo1
|
forkhead box O1
|
|
chr8_+_36740640
|
5.759
|
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr6_-_129572590
|
5.714
|
NM_033078
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr2_+_145647868
|
5.693
|
|
Rin2
|
Ras and Rab interactor 2
|
|
chr2_-_44965603
|
5.589
|
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_163462584
|
5.579
|
|
Serinc3
|
serine incorporator 3
|
|
chr2_+_158157639
|
5.558
|
|
Lbp
|
lipopolysaccharide binding protein
|
|
chr1_-_52714729
|
5.510
|
|
Mfsd6
|
major facilitator superfamily domain containing 6
|
|
chr7_-_110962324
|
5.504
|
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr1_+_19202046
|
5.316
|
NM_001025305
|
Tcfap2b
|
transcription factor AP-2 beta
|
|
chr5_-_17341512
|
5.180
|
|
Cd36
|
CD36 antigen
|
|
chr1_+_59543450
|
5.102
|
|
|
|
|
chr1_-_45460310
|
5.063
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr3_-_20142068
|
5.041
|
NM_007753
|
Cpa3
|
carboxypeptidase A3, mast cell
|
|
chr15_+_3220766
|
5.034
|
NM_001042613
NM_001042614
NM_009155
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr15_+_3220994
|
4.996
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr12_-_104681842
|
4.901
|
NM_029803
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A
|
|
chr5_+_88431549
|
4.894
|
NM_011422
|
Smr3a
|
submaxillary gland androgen regulated protein 3A
|
|
chr1_+_45401303
|
4.893
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr19_+_25073995
|
4.849
|
NM_028785
|
Dock8
|
dedicator of cytokinesis 8
|
|
chr19_-_41289045
|
4.841
|
|
Tm9sf3
|
transmembrane 9 superfamily member 3
|
|
chr19_+_7631935
|
4.838
|
NM_139269
|
Pla2g16
|
phospholipase A2, group XVI
|
|
chr15_-_42508294
|
4.713
|
NM_009640
|
Angpt1
|
angiopoietin 1
|
|
chr6_-_69494873
|
4.710
|
|
Igkv4-57-1
|
immunoglobulin kappa variable 4-57-1
|
|
chr19_-_41068625
|
4.649
|
|
Blnk
|
B-cell linker
|
|
chr1_-_75132608
|
4.646
|
|
1810031K17Rik
|
RIKEN cDNA 1810031K17 gene
|
|
chr19_+_11518042
|
4.601
|
NM_021718
|
Ms4a4b
|
membrane-spanning 4-domains, subfamily A, member 4B
|
|
chr15_+_3220999
|
4.573
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr6_-_130287518
|
4.531
|
NM_010648
|
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3
|
|
chr1_-_55044032
|
4.471
|
|
Sf3b1
|
splicing factor 3b, subunit 1
|
|
chr7_-_110962416
|
4.421
|
NM_008220
NM_016956
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr1_-_173579057
|
4.407
|
|
Slamf7
|
SLAM family member 7
|
|
chr14_-_55725370
|
4.385
|
NM_007455
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit
|
|
chr5_+_104728299
|
4.376
|
NM_008318
|
Ibsp
|
integrin binding sialoprotein
|
|
chr16_+_48872720
|
4.309
|
NM_181596
|
Retnlg
|
resistin like gamma
|
|
chr6_+_70648433
|
4.306
|
|
Igkv3-2
|
immunoglobulin kappa variable 3-2
|
|
chr5_+_88314719
|
4.299
|
NM_027128
|
Odam
|
odontogenic, ameloblast asssociated
|
|
chr9_-_110959702
|
4.289
|
NM_017466
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr4_+_102242664
|
4.264
|
NM_001177980
|
Pde4b
|
phosphodiesterase 4B, cAMP specific
|
|
chr1_-_173537404
|
4.230
|
NM_008534
|
Ly9
|
lymphocyte antigen 9
|
|
chr11_+_67013553
|
4.213
|
NM_030679
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult
|
|
chr6_+_41797547
|
4.209
|
NM_008843
|
Pip
|
prolactin induced protein
|
|
chr5_-_17341713
|
4.207
|
NM_001159555
NM_007643
|
Cd36
|
CD36 antigen
|
|
chr7_+_25682507
|
4.202
|
NM_007655
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha)
|
|
chr9_-_70452528
|
4.144
|
|
Fam63b
|
family with sequence similarity 63, member B
|
|
chr1_-_196957466
|
4.136
|
|
Cr1l
|
complement component (3b/4b) receptor 1-like
|
|
chr13_-_102453449
|
4.128
|
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)
|
|
chr16_+_44820840
|
4.047
|
NM_207244
|
Cd200r4
|
CD200 receptor 4
|
|
chr15_+_102289600
|
4.019
|
NM_001170911
NM_025385
|
Prr13
|
proline rich 13
|
|
chr6_+_38868942
|
4.019
|
NM_011539
|
Tbxas1
|
thromboxane A synthase 1, platelet
|
|
chr7_+_104599843
|
4.015
|
NM_001136235
|
Kctd14
|
potassium channel tetramerisation domain containing 14
|
|
chr14_-_64035873
|
4.014
|
NM_007549
|
Blk
|
B lymphoid kinase
|
|
chr10_-_18731766
|
3.957
|
NM_001166402
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr11_+_67051530
|
3.938
|
NM_010855
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle
|
|
chr9_-_110959315
|
3.897
|
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr6_+_129528268
|
3.837
|
NM_153590
|
Klre1
|
killer cell lectin-like receptor family E member 1
|
|
chr11_+_83476085
|
3.820
|
NM_013652
|
Ccl4
|
chemokine (C-C motif) ligand 4
|
|
chr15_-_9459612
|
3.816
|
NM_008372
|
Il7r
|
interleukin 7 receptor
|
|
chr17_+_34400152
|
3.777
|
NM_207105
|
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1
|
|
chr12_-_115170216
|
3.776
|
|
Ighg
|
Immunoglobulin heavy chain (gamma polypeptide)
|
|
chr1_-_66981900
|
3.742
|
NM_001113387
|
Myl1
|
myosin, light polypeptide 1
|
|
chr6_-_129422702
|
3.742
|
NM_020008
|
Clec7a
|
C-type lectin domain family 7, member a
|
|
chr6_-_136806041
|
3.724
|
NM_026639
|
Art4
|
ADP-ribosyltransferase 4
|
|
chr2_-_60589953
|
3.715
|
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr11_-_79337178
|
3.704
|
|
Evi2b
|
ecotropic viral integration site 2b
|
|
chr6_-_124586102
|
3.692
|
NM_173864
|
Gm5077
|
predicted gene 5077
|
|
chr4_-_44053229
|
3.665
|
|
Gne
|
glucosamine
|
|
chr1_+_87546558
|
3.660
|
NM_013673
|
Sp100
|
nuclear antigen Sp100
|
|
chr3_+_51299646
|
3.643
|
|
|
|
|
chr10_+_88348725
|
3.626
|
NM_145423
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8
|
|
chr17_-_40379205
|
3.600
|
NM_009639
|
Crisp3
|
cysteine-rich secretory protein 3
|
|
chr1_-_146622545
|
3.574
|
NM_022881
|
Rgs18
|
regulator of G-protein signaling 18
|
|
chr15_-_5058532
|
3.567
|
NM_001163138
|
Card6
|
caspase recruitment domain family, member 6
|
|
chr1_-_146024404
|
3.555
|
NM_153171
|
Rgs13
|
regulator of G-protein signaling 13
|
|
chr6_+_70165393
|
3.546
|
|
Igk-V28
Igkv6-23
LOC672450
|
immunoglobulin kappa chain variable 28 (V28)
immunoglobulin kappa variable 6-23
V(kappa) gene product
|
|
chr1_+_109407998
|
3.520
|
NM_001174170
NM_011111
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2
|
|
chr10_+_96945972
|
3.487
|
|
Dcn
|
decorin
|
|
chr10_-_28706096
|
3.465
|
NM_026336
|
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene
|
|
chr9_+_5308846
|
3.462
|
NM_007609
|
Casp4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr9_+_15309938
|
3.387
|
NM_133214
|
BC017612
|
cDNA sequence BC017612
|
|
chr7_+_27904968
|
3.385
|
NM_007817
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr2_-_59963796
|
3.373
|
NM_001001182
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr6_-_130183339
|
3.341
|
NM_001110323
|
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7
|
|
chr18_+_20469364
|
3.326
|
NM_010079
|
Dsg1a
|
desmoglein 1 alpha
|
|
chr2_-_44965866
|
3.302
|
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr17_+_47641999
|
3.271
|
NM_001081635
NM_001081636
|
Ccnd3
|
cyclin D3
|
|
chr1_+_7162595
|
3.268
|
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr2_+_112106470
|
3.256
|
NM_133649
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr2_+_110561733
|
3.234
|
NM_172979
|
Muc15
|
mucin 15
|
|
chr5_-_98459943
|
3.233
|
NM_133738
|
Antxr2
|
anthrax toxin receptor 2
|
|
chr9_+_45127182
|
3.226
|
NM_001013373
|
Tmprss13
|
transmembrane protease, serine 13
|
|
chr2_+_145648290
|
3.214
|
|
Rin2
|
Ras and Rab interactor 2
|
|
chr11_-_79343891
|
3.199
|
|
Evi2a
|
ecotropic viral integration site 2a
|
|
chr7_+_27904997
|
3.171
|
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr4_+_94405963
|
3.139
|
NM_013690
|
Tek
|
endothelial-specific receptor tyrosine kinase
|
|
chr17_-_51965912
|
3.128
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr1_+_109258889
|
3.120
|
NM_025867
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11
|
|
chr11_-_79344055
|
3.117
|
NM_001033711
NM_010161
NM_146023
|
Evi2a
Evi2b
|
ecotropic viral integration site 2a
ecotropic viral integration site 2b
|
|
chr16_+_49913305
|
3.116
|
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer)
|
|
chr6_+_67966702
|
3.111
|
|
|
|
|
chr5_-_88128419
|
3.108
|
NM_009972
|
Csn2
|
casein beta
|
|
chr7_+_27904981
|
3.105
|
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr15_-_76853069
|
3.046
|
NM_001164048
NM_001164047
|
Mb
|
myoglobin
|
|
chr4_+_94406190
|
3.044
|
|
Tek
|
endothelial-specific receptor tyrosine kinase
|
|
chr10_-_83989221
|
3.039
|
|
Ckap4
|
cytoskeleton-associated protein 4
|
|
chr13_+_118009379
|
3.038
|
NM_010330
|
Emb
|
embigin
|
|
chr18_+_11815869
|
3.025
|
|
Rbbp8
|
retinoblastoma binding protein 8
|
|
chrX_-_150130711
|
3.024
|
NM_175429
|
Kctd12b
|
potassium channel tetramerisation domain containing 12b
|
|
chr1_-_196957694
|
3.001
|
NM_013499
|
Cr1l
|
complement component (3b/4b) receptor 1-like
|
|
chr3_+_93324437
|
2.995
|
|
S100a11
|
S100 calcium binding protein A11 (calgizzarin)
|
|
chr1_+_154622471
|
2.952
|
|
|
|
|
chr11_-_106576464
|
2.949
|
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr6_-_142271469
|
2.936
|
NM_130861
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5
|
|
chr8_+_90806890
|
2.935
|
NM_001109756
|
Adcy7
|
adenylate cyclase 7
|
|
chr11_-_106576585
|
2.918
|
NM_001032378
NM_008816
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr5_+_123093892
|
2.916
|
NM_001038839
NM_001038845
NM_001038887
NM_011027
|
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr6_-_113385745
|
2.911
|
NM_178373
|
Cidec
|
cell death-inducing DFFA-like effector c
|
|
chr5_-_65351226
|
2.904
|
NM_011604
|
Tlr6
|
toll-like receptor 6
|
|
chr10_-_117252126
|
2.894
|
|
|
|
|
chr3_-_20022409
|
2.894
|
|
Gyg
|
glycogenin
|
|
chr11_-_106576380
|
2.893
|
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr3_-_58934492
|
2.888
|
NM_001008497
NM_133200
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr6_+_70321430
|
2.881
|
|
|
|
|
chr16_+_48872756
|
2.881
|
|
Retnlg
|
resistin like gamma
|
|
chr11_-_62206116
|
2.862
|
|
Ncor1
|
nuclear receptor co-repressor 1
|
|
chr14_+_27813547
|
2.861
|
NM_010420
|
Hesx1
|
homeobox gene expressed in ES cells
|
|
chr4_-_108790523
|
2.857
|
|
Osbpl9
|
oxysterol binding protein-like 9
|
|
chr19_-_41289262
|
2.841
|
|
Tm9sf3
|
transmembrane 9 superfamily member 3
|
|
chr16_+_49868005
|
2.840
|
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer)
|
|
chr1_-_164816645
|
2.839
|
|
Fmo2
|
flavin containing monooxygenase 2
|
|
chr6_-_115712483
|
2.836
|
NM_001168256
NM_001168257
|
Tmem40
|
transmembrane protein 40
|
|
chr1_+_173612132
|
2.824
|
|
Cd48
|
CD48 antigen
|
|
chr4_-_118862555
|
2.807
|
NM_013848
|
Ermap
|
erythroblast membrane-associated protein
|
|
chr3_+_93324411
|
2.792
|
NM_016740
|
S100a11
|
S100 calcium binding protein A11 (calgizzarin)
|
|
chr19_+_12535268
|
2.792
|
NM_010821
|
Mpeg1
|
macrophage expressed gene 1
|
|
chr6_-_122799694
|
2.788
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr5_+_148242018
|
2.751
|
NM_028291
|
Pan3
|
PAN3 polyA specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr9_+_7272513
|
2.749
|
NM_008607
|
Mmp13
|
matrix metallopeptidase 13
|
|
chr2_-_66512442
|
2.745
|
|
Scn7a
|
sodium channel, voltage-gated, type VII, alpha
|
|
chr3_+_67406958
|
2.730
|
|
Mfsd1
|
major facilitator superfamily domain containing 1
|
|
chr11_-_46202942
|
2.723
|
NM_010583
|
Itk
|
IL2-inducible T-cell kinase
|
|
chr2_-_119916315
|
2.709
|
|
Ehd4
|
EH-domain containing 4
|
|
chr17_-_34960292
|
2.685
|
NM_011413
|
C4a
|
complement component 4A (Rodgers blood group)
|
|
chr8_+_131028275
|
2.684
|
|
Nrp1
|
neuropilin 1
|
|
chr19_-_20802015
|
2.679
|
NM_011921
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7
|
|
chr6_-_3349470
|
2.675
|
NM_010156
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr11_-_106576550
|
2.666
|
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr17_-_40456121
|
2.653
|
NM_009638
|
Crisp1
|
cysteine-rich secretory protein 1
|
|
chr16_+_43640198
|
2.650
|
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_35331446
|
2.632
|
NM_008518
|
Ltb
|
lymphotoxin B
|
|
chr4_+_66488753
|
2.623
|
NM_021297
|
Tlr4
|
toll-like receptor 4
|