|
chr4_-_131978279
|
6.241
|
NM_001161797
NM_175306
|
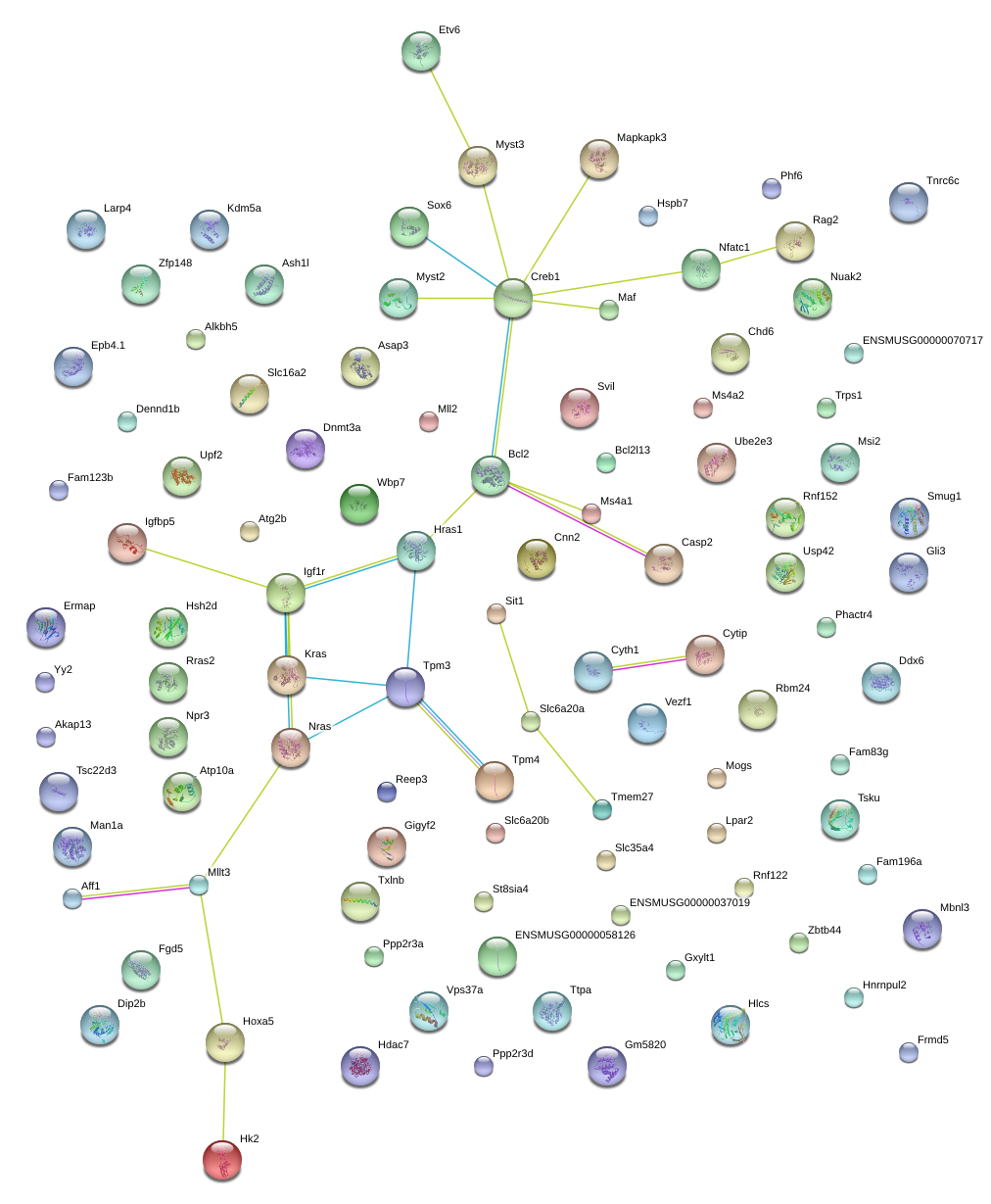
Phactr4
|
phosphatase and actin regulator 4
|
|
chr11_-_88579517
|
4.099
|
NM_054043
|
Msi2
|
Musashi homolog 2 (Drosophila)
|
|
chr3_+_89876566
|
4.042
|
NM_022314
|
Tpm3
|
tropomyosin 3, gamma
|
|
chr1_-_72921438
|
3.907
|
NM_010518
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr6_+_120786207
|
3.542
|
NM_153516
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator)
|
|
chr7_+_65913571
|
3.403
|
NM_009728
|
Atp10a
|
ATPase, class V, type 10A
|
|
chr9_+_30838224
|
3.157
|
NM_001115130
NM_172765
|
Zbtb44
|
zinc finger and BTB domain containing 44
|
|
chr6_+_133985717
|
3.034
|
NM_007961
|
Etv6
|
ets variant gene 6 (TEL oncogene)
|
|
chr15_-_11835393
|
2.998
|
NM_001039181
NM_008728
|
Npr3
|
natriuretic peptide receptor 3
|
|
chr18_+_39152776
|
2.936
|
NM_175164
|
Arhgap26
|
Rho GTPase activating protein 26
|
|
chr7_+_82600419
|
2.751
|
NM_029332
|
Akap13
|
A kinase (PRKA) anchor protein 13
|
|
chr5_+_104183148
|
2.687
|
NM_133919
|
Aff1
|
AF4/FMR2 family, member 1
|
|
chr5_+_104183591
|
2.442
|
NM_001080798
|
Aff1
|
AF4/FMR2 family, member 1
|
|
chr1_-_97564139
|
2.366
|
NM_001159745
NM_009183
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr8_+_72346327
|
2.059
|
NM_020028
|
Lpar2
|
lysophosphatidic acid receptor 2
|
|
chr4_-_87679175
|
2.048
|
NM_027326
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr13_+_46513668
|
1.898
|
NM_001081425
|
Rbm24
|
RNA binding motif protein 24
|
|
chr11_-_95171521
|
1.786
|
NM_001195003
NM_001195004
NM_177619
|
Myst2
|
MYST histone acetyltransferase 2
|
|
chr15_-_50721552
|
1.768
|
NM_032000
|
Trps1
|
trichorhinophalangeal syndrome I (human)
|
|
chr6_+_83065466
|
1.736
|
NM_020619
|
Mogs
|
mannosyl-oligosaccharide glucosidase
|
|
chr9_-_107192203
|
1.717
|
NM_178907
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chrX_-_101017286
|
1.687
|
NM_009197
|
Slc16a2
|
solute carrier family 16 (monocarboxylic acid transporters), member 2
|
|
chr18_+_5046584
|
1.686
|
NM_153153
|
Svil
|
supervillin
|
|
chr8_-_118230771
|
1.685
|
NM_001025577
|
Maf
|
avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog
|
|
chr4_+_135762279
|
1.628
|
NM_001008232
|
Asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr6_-_82724366
|
1.608
|
NM_013820
|
Hk2
|
hexokinase 2
|
|
chr2_-_58012447
|
1.596
|
NM_139200
|
Cytip
|
cytohesin 1 interacting protein
|
|
chr1_+_134212642
|
1.591
|
NM_001195025
NM_028778
|
Nuak2
|
NUAK family, SNF1-like kinase, 2
|
|
chr12_+_3806955
|
1.566
|
NM_007872
|
Dnmt3a
|
DNA methyltransferase 3A
|
|
chr16_+_33380860
|
1.516
|
NM_011749
|
Zfp148
|
zinc finger protein 148
|
|
chr10_-_53795556
|
1.443
|
NM_008548
|
Man1a
|
mannosidase 1, alpha
|
|
chrX_-_137077639
|
1.400
|
NM_010286
|
Tsc22d3
|
TSC22 domain family, member 3
|
|
chr10_+_17516024
|
1.316
|
NM_138628
|
Txlnb
|
taxilin beta
|
|
chr2_-_160934743
|
1.311
|
NM_173368
|
Chd6
|
chromodomain helicase DNA binding protein 6
|
|
chr7_-_123182257
|
1.213
|
NM_001025559
|
Sox6
|
SRY-box containing gene 6
|
|
chr6_-_145198693
|
1.188
|
NM_021284
|
Kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
|
|
chr16_-_94509462
|
1.179
|
NM_139145
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase)
|
|
chr9_+_44412931
|
1.175
|
NM_001110826
NM_007841
NM_181324
|
Ddx6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr4_-_131631226
|
1.174
|
NM_001128607
|
Epb4.1
|
erythrocyte protein band 4.1
|
|
chr15_-_93105514
|
1.120
|
NM_001033275
|
Gxylt1
|
glucoside xylosyltransferase 1
|
|
chr7_+_75097142
|
1.106
|
NM_010513
|
Igf1r
|
insulin-like growth factor I receptor
|
|
chr2_-_121632749
|
1.078
|
NM_172673
|
Frmd5
|
FERM domain containing 5
|
|
chr15_+_99803210
|
1.076
|
NM_001024526
|
Larp4
|
La ribonucleoprotein domain family, member 4
|
|
chr3_+_88769733
|
1.073
|
NM_138679
|
Ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr4_+_140976693
|
1.069
|
NM_013868
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr15_-_98701613
|
1.066
|
NM_001033276
|
Mll2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr4_-_131604821
|
1.062
|
NM_001128606
|
Epb4.1
|
erythrocyte protein band 4.1
|
|
chr13_+_15555483
|
1.055
|
NM_008130
|
Gli3
|
GLI-Kruppel family member GLI3
|
|
chr11_+_60351159
|
1.044
|
NM_172943
|
Alkbh5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr1_-_107253245
|
0.990
|
NM_001160368
NM_178779
|
Rnf152
|
ring finger protein 152
|
|
chr1_+_140860285
|
0.989
|
NM_001166501
|
Dennd1b
|
DENN/MADD domain containing 1B
|
|
chr10_-_66559568
|
0.979
|
NM_178606
|
Reep3
|
receptor accessory protein 3
|
|
chr11_+_117515548
|
0.974
|
NM_198022
|
Tnrc6c
|
trinucleotide repeat containing 6C
|
|
chrX_-_154036606
|
0.962
|
NM_172307
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr4_-_43496556
|
0.961
|
NM_019436
|
Sit1
|
suppression inducing transmembrane adaptor 1
|
|
chrX_+_160528118
|
0.948
|
NM_020626
|
Tmem27
|
transmembrane protein 27
|
|
chr15_-_97662101
|
0.923
|
NM_019572
|
Hdac7
|
histone deacetylase 7
|
|
chr7_-_142129892
|
0.917
|
NM_001143802
|
Fam196a
|
family with sequence similarity 196, member A
|
|
chrX_-_92640149
|
0.907
|
NM_175179
|
Fam123b
|
family with sequence similarity 123, member B
|
|
chr11_+_61497911
|
0.905
|
NM_178618
|
Fam83g
|
family with sequence similarity 83, member G
|
|
chrX_+_50265390
|
0.902
|
NM_027642
|
Phf6
|
PHD finger protein 6
|
|
chr15_+_99869071
|
0.897
|
NM_001159361
|
Dip2b
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chrX_-_48559008
|
0.887
|
NM_134163
|
Mbnl3
|
muscleblind-like 3 (Drosophila)
|
|
chr8_+_23970006
|
0.886
|
NM_001081149
|
Myst3
|
MYST histone acetyltransferase (monocytic leukemia) 3
|
|
chr7_-_121261292
|
0.880
|
NM_025846
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2
|
|
chr18_-_80909796
|
0.878
|
NM_001164109
NM_001164110
NM_198429
|
Nfatc1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr4_-_87452140
|
0.860
|
NM_029931
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr19_-_11340591
|
0.842
|
NM_007641
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr7_-_105509797
|
0.839
|
NM_001024619
NM_001168540
NM_001168541
NM_001168539
|
Tsku
|
tsukushin
|
|
chr8_+_74659107
|
0.839
|
NM_001001491
|
Tpm4
|
tropomyosin 4
|
|
chr18_-_80904852
|
0.838
|
NM_001164111
NM_001164112
NM_016791
|
Nfatc1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr1_+_89223558
|
0.830
|
NM_146112
NM_001110212
|
Gigyf2
|
GRB10 interacting GYF protein 2
|
|
chr6_+_91937103
|
0.811
|
NM_172731
|
Fgd5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr1_-_108610861
|
0.810
|
NM_009741
NM_177410
|
Bcl2
|
B-cell leukemia/lymphoma 2
|
|
chr6_+_42214999
|
0.791
|
NM_007610
|
Casp2
|
caspase 2
|
|
chr1_+_64579358
|
0.788
|
NM_001037726
NM_009952
NM_133828
|
Creb1
|
cAMP responsive element binding protein 1
|
|
chr9_-_101154160
|
0.783
|
NM_001161362
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr8_+_32222291
|
0.779
|
NM_175136
|
Rnf122
|
ring finger protein 122
|
|
chr4_+_19935558
|
0.771
|
NM_015767
|
Ttpa
|
tocopherol (alpha) transfer protein
|
|
chr5_-_144493077
|
0.744
|
NM_029749
|
Usp42
|
ubiquitin specific peptidase 42
|
|
chr7_-_123174560
|
0.742
|
NM_001025560
|
Sox6
|
SRY-box containing gene 6
|
|
chr18_+_36838868
|
0.732
|
NM_001083317
NM_026404
|
Slc35a4
|
solute carrier family 35, member A4
|
|
chr10_+_79451332
|
0.717
|
NM_007725
|
Cnn2
|
calponin 2
|
|
chr2_+_78709195
|
0.687
|
NM_009454
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3, UBC4/5 homolog (yeast)
|
|
chr11_+_87881747
|
0.683
|
NM_016686
|
Vezf1
|
vascular endothelial zinc finger 1
|
|
chr8_+_74713536
|
0.681
|
NM_197944
|
Hsh2d
|
hematopoietic SH2 domain containing
|
|
chr6_-_52154455
|
0.679
|
NM_010453
|
Hoxa5
|
homeobox A5
|
|
chr8_+_41597112
|
0.654
|
NM_033560
|
Vps37a
|
vacuolar protein sorting 37A (yeast)
|
|
chr6_+_120314116
|
0.653
|
NM_145997
|
Kdm5a
|
lysine (K)-specific demethylase 5A
|
|
chr4_-_118862555
|
0.651
|
NM_013848
|
Ermap
|
erythroblast membrane-associated protein
|
|
chr15_-_102993714
|
0.646
|
NM_027885
|
Smug1
|
single-strand selective monofunctional uracil DNA glycosylase
|
|
chr2_+_5872500
|
0.643
|
NM_001081132
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr1_+_140864297
|
0.641
|
NM_181347
|
Dennd1b
|
DENN/MADD domain containing 1B
|
|
chr2_+_101464874
|
0.622
|
NM_009020
|
Rag2
|
recombination activating gene 2
|
|
chr12_-_106923394
|
0.617
|
NM_029654
|
Atg2b
|
ATG2 autophagy related 2 homolog B (S. cerevisiae)
|
|
chr19_-_47538898
|
0.616
|
NM_001164717
NM_008018
|
Sh3pxd2a
|
SH3 and PX domains 2A
|
|
chr13_-_58712170
|
0.613
|
NM_022317
|
Slc28a3
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 3
|
|
chr19_+_46647844
|
0.610
|
NM_053196
|
Sfxn2
|
sideroflexin 2
|
|
chr6_-_71390592
|
0.610
|
NM_024288
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr7_-_123138599
|
0.608
|
NM_011445
|
Sox6
|
SRY-box containing gene 6
|
|
chr6_+_100783598
|
0.604
|
NM_182939
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr8_-_125201971
|
0.598
|
NM_001109873
NM_177289
|
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human)
|
|
chr15_-_96529998
|
0.593
|
NM_175121
|
Slc38a2
|
solute carrier family 38, member 2
|
|
chr4_+_48137778
|
0.587
|
NM_026343
|
Stx17
|
syntaxin 17
|
|
chr4_+_21703416
|
0.583
|
NM_152825
|
Usp45
|
ubiquitin specific petidase 45
|
|
chr15_+_102797295
|
0.574
|
NM_010462
|
Hoxc10
|
homeobox C10
|
|
chr11_+_95198329
|
0.569
|
NM_172543
|
Fam117a
|
family with sequence similarity 117, memberA
|
|
chr19_-_42827264
|
0.567
|
NM_029011
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2
|
|
chr11_+_98064618
|
0.566
|
NM_001109626
NM_001109628
NM_026952
|
Cdk12
|
cyclin-dependent kinase 12
|
|
chr10_+_120802099
|
0.561
|
NM_029364
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr5_-_63157309
|
0.559
|
NM_178407
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr12_-_86680251
|
0.555
|
NM_145138
|
Nek9
|
NIMA (never in mitosis gene a)-related expressed kinase 9
|
|
chr18_-_39078304
|
0.553
|
NM_010197
|
Fgf1
|
fibroblast growth factor 1
|
|
chr8_-_127473082
|
0.549
|
NM_053207
|
Egln1
|
EGL nine homolog 1 (C. elegans)
|
|
chr13_-_37141055
|
0.547
|
NM_028784
|
F13a1
|
coagulation factor XIII, A1 subunit
|
|
chr1_-_153937364
|
0.543
|
NM_197990
|
LOC545371
1700025G04Rik
|
hypothetical protein LOC545371
RIKEN cDNA 1700025G04 gene
|
|
chr11_-_34597324
|
0.541
|
NM_033374
|
Dock2
|
dedicator of cyto-kinesis 2
|
|
chr9_-_119932620
|
0.541
|
NM_011724
|
Xirp1
|
xin actin-binding repeat containing 1
|
|
chr1_-_157820378
|
0.540
|
NM_001039184
|
Cep350
|
centrosomal protein 350
|
|
chr18_+_42435006
|
0.538
|
NM_172626
|
Rbm27
|
RNA binding motif protein 27
|
|
chr1_+_191552133
|
0.538
|
NM_008976
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr9_+_108984968
|
0.534
|
NM_025689
|
Ccdc51
|
coiled-coil domain containing 51
|
|
chr7_+_28964486
|
0.531
|
NM_001037957
NM_010092
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b
|
|
chr7_+_13512764
|
0.529
|
NM_029809
|
2310014L17Rik
|
RIKEN cDNA 2310014L17 gene
|
|
chr15_+_99984826
|
0.529
|
NM_172819
|
Dip2b
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr15_+_99800504
|
0.518
|
NM_001080948
|
Larp4
|
La ribonucleoprotein domain family, member 4
|
|
chr15_+_25772997
|
0.515
|
NM_001034851
|
Fam134b
|
family with sequence similarity 134, member B
|
|
chr4_-_106850812
|
0.514
|
NM_025617
|
2210012G02Rik
|
RIKEN cDNA 2210012G02 gene
|
|
chr16_+_35938555
|
0.508
|
NM_030253
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9
|
|
chr3_+_94928435
|
0.504
|
NM_016712
|
Tmod4
|
tropomodulin 4
|
|
chrX_+_103382432
|
0.499
|
NM_008828
|
Pgk1
|
phosphoglycerate kinase 1
|
|
chr1_-_156573049
|
0.499
|
NM_009782
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr9_-_108092712
|
0.498
|
NM_007567
|
Bsn
|
bassoon
|
|
chr11_+_75007475
|
0.493
|
NM_177708
|
Rtn4rl1
|
reticulon 4 receptor-like 1
|
|
chr4_-_126645129
|
0.491
|
NM_001114399
|
Zmym4
|
zinc finger, MYM-type 4
|
|
chr13_-_37142112
|
0.491
|
NM_001166391
|
F13a1
|
coagulation factor XIII, A1 subunit
|
|
chr8_-_125222837
|
0.490
|
NM_009824
|
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human)
|
|
chr8_+_107800316
|
0.487
|
NM_030152
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr2_-_28771940
|
0.482
|
NM_019446
NM_001164186
|
Barhl1
|
BarH-like 1 (Drosophila)
|
|
chr9_+_64027101
|
0.475
|
NM_183316
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5
|
|
chr14_-_73725576
|
0.473
|
NM_009029
|
Rb1
|
retinoblastoma 1
|
|
chr15_+_37162790
|
0.472
|
NM_026496
|
Grhl2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_-_186635140
|
0.469
|
NM_001081361
|
Mosc1
|
MOCO sulphurase C-terminal domain containing 1
|
|
chr4_-_58219368
|
0.467
|
NM_022814
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr9_-_20619426
|
0.466
|
NM_016919
|
Col5a3
|
collagen, type V, alpha 3
|
|
chr11_-_100711785
|
0.464
|
NM_001113563
|
Stat5b
|
signal transducer and activator of transcription 5B
|
|
chr9_+_69245718
|
0.463
|
NM_145618
|
Narg2
|
NMDA receptor-regulated gene 2
|
|
chr4_-_131628011
|
0.459
|
NM_183428
|
Epb4.1
|
erythrocyte protein band 4.1
|
|
chr12_+_113917038
|
0.453
|
NM_001100460
|
Zbtb42
|
zinc finger and BTB domain containing 42
|
|
chr1_-_157589156
|
0.452
|
NM_010712
|
Lhx4
|
LIM homeobox protein 4
|
|
chr14_+_52604434
|
0.451
|
NM_001145921
NM_001145922
NM_198249
|
Arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr18_-_82862111
|
0.451
|
NM_177832
|
Zfp236
|
zinc finger protein 236
|
|
chr3_+_95908422
|
0.448
|
NM_001025613
NM_001025614
|
Otud7b
|
OTU domain containing 7B
|
|
chrX_+_132758579
|
0.448
|
NM_001127169
|
Tceal7
|
transcription elongation factor A (SII)-like 7
|
|
chr2_+_110561733
|
0.445
|
NM_172979
|
Muc15
|
mucin 15
|
|
chr3_+_97432726
|
0.445
|
NM_001161763
NM_001161765
NM_010232
|
Fmo5
|
flavin containing monooxygenase 5
|
|
chr2_+_179777187
|
0.442
|
NM_178750
|
Ss18l1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr19_+_21727795
|
0.430
|
NM_146096
|
Fam108b
|
family with sequence similarity 108, member B
|
|
chr4_-_70195952
|
0.428
|
NM_172694
|
Megf9
|
multiple EGF-like-domains 9
|
|
chr11_-_106134024
|
0.419
|
NM_001130187
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr7_-_96666081
|
0.418
|
NM_029614
|
Prss23
|
protease, serine, 23
|
|
chr2_+_4638844
|
0.412
|
NM_001190400
NM_178663
|
Bend7
|
BEN domain containing 7
|
|
chr11_-_106128935
|
0.410
|
NM_031878
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr7_+_88202481
|
0.410
|
NM_054085
|
Alpk3
|
alpha-kinase 3
|
|
chr3_+_68494565
|
0.407
|
NM_001159424
|
Il12a
|
interleukin 12a
|
|
chr2_-_93303102
|
0.403
|
NM_001136055
|
Cd82
|
CD82 antigen
|
|
chr8_+_107694536
|
0.402
|
NM_001161456
NM_001161457
NM_001161458
NM_022309
|
Cbfb
|
core binding factor beta
|
|
chr3_-_95054970
|
0.400
|
NM_001168356
NM_134253
|
Bnipl
|
BCL2/adenovirus E1B 19kD interacting protein like
|
|
chr5_-_9161670
|
0.400
|
NM_001110327
NM_011806
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1
|
|
chr17_+_87682206
|
0.397
|
NM_028639
|
Ttc7
|
tetratricopeptide repeat domain 7
|
|
chr8_-_108457731
|
0.397
|
NM_023182
|
Ctrl
|
chymotrypsin-like
|
|
chr15_+_102236709
|
0.396
|
NM_013672
|
Sp1
|
trans-acting transcription factor 1
|
|
chr19_+_11986887
|
0.395
|
NM_172635
|
Patl1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr1_+_36568707
|
0.393
|
NM_001039551
NM_053186
|
Cnnm3
|
cyclin M3
|
|
chr2_+_150043834
|
0.393
|
NM_001142411
|
Zfp937
|
zinc finger protein 937
|
|
chr13_-_94269616
|
0.392
|
NM_021310
|
Jmy
|
junction-mediating and regulatory protein
|
|
chr3_-_95021614
|
0.391
|
NM_029885
NM_172512
|
Gabpb2
|
GA repeat binding protein, beta 2
|
|
chr4_+_41082993
|
0.390
|
NM_026275
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr3_+_68495345
|
0.389
|
NM_008351
|
Il12a
|
interleukin 12a
|
|
chr6_-_148894953
|
0.386
|
NM_019643
|
Fam60a
|
family with sequence similarity 60, member A
|
|
chr3_+_122432075
|
0.384
|
NM_153422
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific
|
|
chr6_-_119494295
|
0.384
|
NM_009525
|
Wnt5b
|
wingless-related MMTV integration site 5B
|
|
chr9_+_53345151
|
0.379
|
NM_001081152
|
Npat
|
nuclear protein in the AT region
|
|
chr9_-_70504934
|
0.379
|
NM_172772
|
Fam63b
|
family with sequence similarity 63, member B
|
|
chr6_+_71272683
|
0.378
|
NM_009858
|
Cd8b1
|
CD8 antigen, beta chain 1
|
|
chr3_+_55586431
|
0.375
|
NM_010750
|
Mab21l1
|
mab-21-like 1 (C. elegans)
|
|
chr6_-_58974400
|
0.372
|
NM_153574
|
Fam13a
|
family with sequence similarity 13, member A
|
|
chr16_+_97578333
|
0.371
|
NM_019517
|
Bace2
|
beta-site APP-cleaving enzyme 2
|
|
chr12_+_29360060
|
0.370
|
NM_134052
|
Adi1
|
acireductone dioxygenase 1
|
|
chr2_-_93302621
|
0.369
|
NM_007656
|
Cd82
|
CD82 antigen
|
|
chr9_-_22112081
|
0.365
|
NM_145612
|
Zfp810
|
zinc finger protein 810
|
|
chr4_-_57314708
|
0.363
|
NM_011207
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr17_+_48594225
|
0.363
|
NM_152823
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like
|
|
chrX_+_68468175
|
0.361
|
NM_001164191
NM_001164192
NM_001164193
NM_019926
|
Mtm1
|
X-linked myotubular myopathy gene 1
|
|
chrX_+_68463941
|
0.355
|
NM_001164190
|
Mtm1
|
X-linked myotubular myopathy gene 1
|
|
chr17_-_10512217
|
0.354
|
NM_001159516
NM_001159517
NM_021881
|
Qk
|
quaking
|
|
chr14_+_49791899
|
0.351
|
NM_001081430
|
Naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit
|
|
chr14_-_68551487
|
0.351
|
NM_177780
|
Dock5
|
dedicator of cytokinesis 5
|
|
chr13_-_23461025
|
0.351
|
NM_001111107
NM_172586
|
Zfp322a
|
zinc finger protein 322A
|
|
chr11_+_76759157
|
0.350
|
NM_178645
|
Blmh
|
bleomycin hydrolase
|