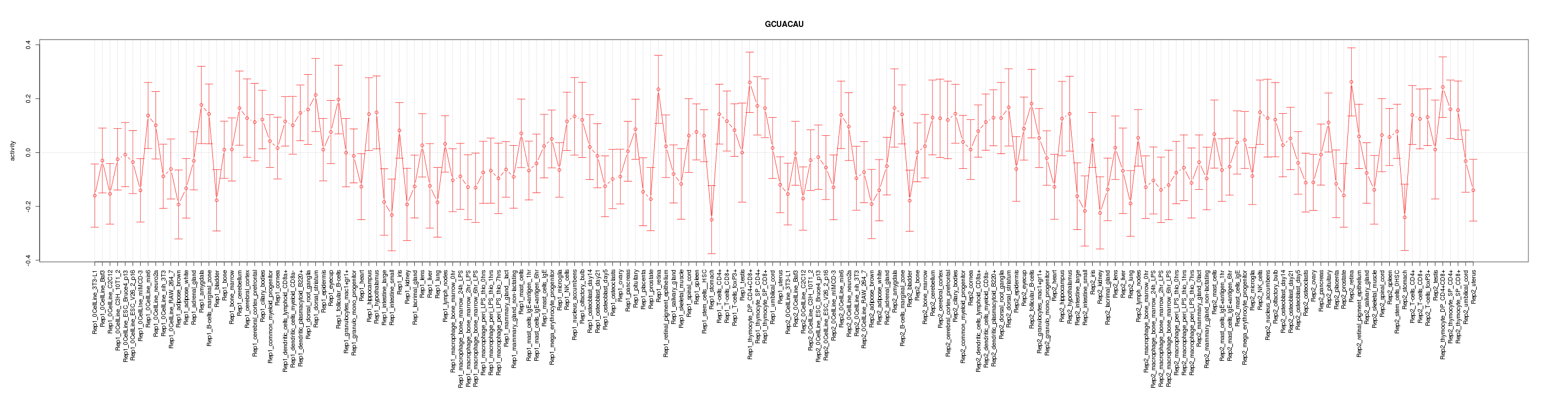
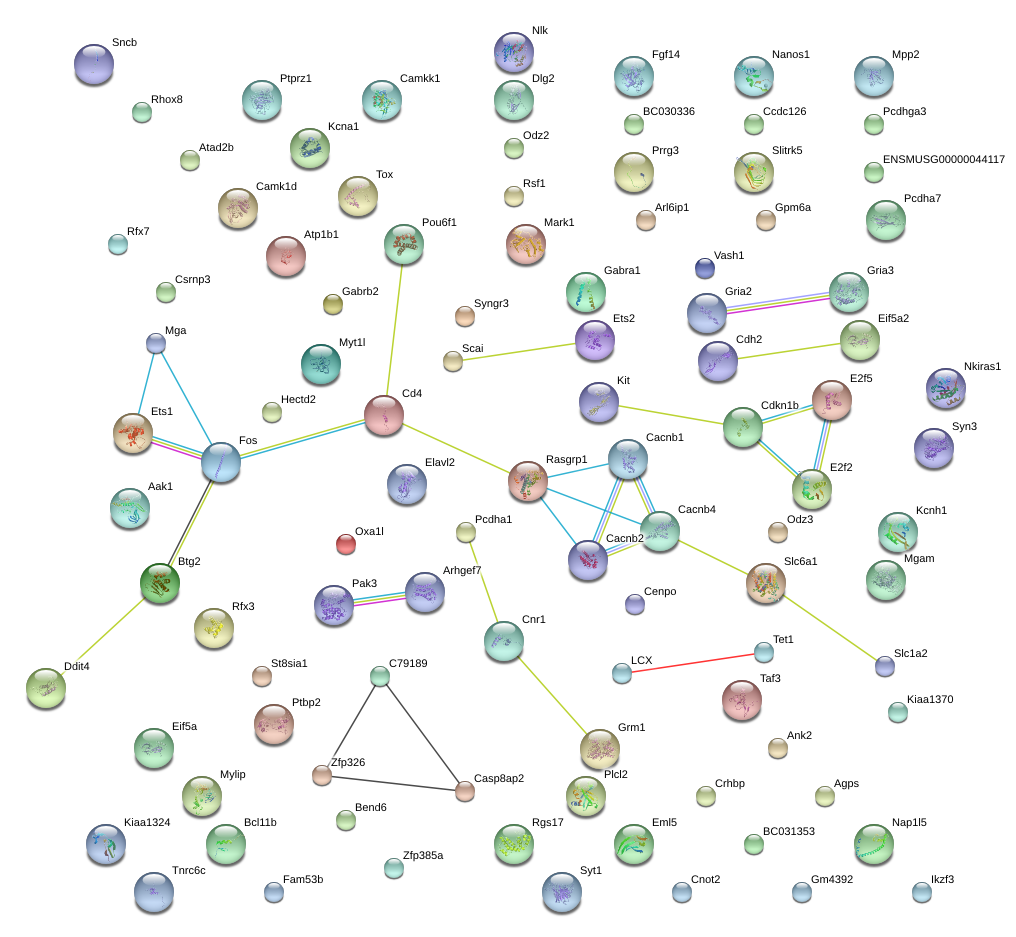
|
chr10_-_62342761
|
12.771
|
NM_027384
|
Tet1
|
tet oncogene 1
|
|
chr18_+_37089837
|
12.622
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chr11_-_41996141
|
12.427
|
NM_010250
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1
|
|
chr18_+_37120093
|
12.021
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr18_+_37127284
|
11.921
|
NM_007767
|
Pcdha6
|
protocadherin alpha 6
|
|
chr18_+_37179802
|
11.652
|
NM_138663
|
Pcdha12
|
protocadherin alpha 12
|
|
chr18_+_37105758
|
11.652
|
NM_138662
|
Pcdha3
|
protocadherin alpha 3
|
|
chr18_+_37303622
|
11.464
|
NM_001003672
|
Pcdhac2
|
protocadherin alpha subfamily C, 2
|
|
chr18_+_37098858
|
11.029
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr18_+_37152024
|
11.013
|
NM_201243
|
Pcdha8
|
protocadherin alpha 8
|
|
chr18_+_37157533
|
10.637
|
NM_138661
|
Pcdha9
|
protocadherin alpha 9
|
|
chr18_+_37133455
|
9.337
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chr10_+_4424140
|
8.757
|
NM_001161822
NM_019958
|
Rgs17
|
regulator of G-protein signaling 17
|
|
chr2_-_5635547
|
8.686
|
NM_177343
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr16_+_13986691
|
8.595
|
NM_144518
|
2900011O08Rik
|
RIKEN cDNA 2900011O08 gene
|
|
chr6_-_124838150
|
8.568
|
NM_013488
|
Cd4
|
CD4 antigen
|
|
chr12_+_86814822
|
7.746
|
NM_010234
|
Fos
|
FBJ osteosarcoma oncogene
|
|
chr2_-_52532100
|
7.555
|
NM_001037099
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chrX_+_69208336
|
7.552
|
NM_001081135
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr9_+_32503600
|
7.545
|
NM_001038642
NM_011808
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain
|
|
chr6_-_142912908
|
7.416
|
NM_011374
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr8_+_56040080
|
6.974
|
NM_153581
|
Gpm6a
|
glycoprotein m6a
|
|
chr2_-_52414072
|
6.944
|
NM_146123
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr18_+_37112301
|
6.634
|
NM_001174154
NM_007766
|
Pcdha4-g
Pcdha4
|
protocadherin alpha 4-gamma
protocadherin alpha 4
|
|
chr9_+_72380046
|
6.417
|
NM_001033536
|
Rfx7
|
regulatory factor X, 7
|
|
chrX_+_140127827
|
6.177
|
NM_001195046
NM_001195047
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr14_+_112074336
|
6.049
|
NM_198865
|
Slitrk5
|
SLIT and NTRK-like family, member 5
|
|
chr6_+_22825464
|
6.044
|
NM_001081306
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1
|
|
chr3_-_119486305
|
5.965
|
NM_019550
|
Ptbp2
|
polypyrimidine tract binding protein 2
|
|
chr3_+_28680223
|
5.691
|
NM_177586
|
Eif5a2
|
eukaryotic translation initiation factor 5A2
|
|
chr8_+_11728104
|
5.510
|
NM_001113517
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7)
|
|
chr4_-_91038729
|
5.379
|
NM_010486
NM_207686
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr12_-_100139666
|
5.266
|
NM_001081191
|
Eml5
|
echinoderm microtubule associated protein like 5
|
|
chr4_-_91066674
|
5.237
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_-_91042997
|
5.175
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr6_+_114232618
|
5.107
|
NM_178703
|
Slc6a1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr2_+_102498839
|
5.101
|
NM_001077514
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chrX_+_139997806
|
5.101
|
NM_008778
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr12_-_109241423
|
5.061
|
NM_001079883
NM_021399
|
Bcl11b
|
B-cell leukemia/lymphoma 11B
|
|
chr11_+_117515548
|
4.828
|
NM_198022
|
Tnrc6c
|
trinucleotide repeat containing 6C
|
|
chr13_-_96214609
|
4.783
|
NM_198408
|
Crhbp
|
corticotropin releasing hormone binding protein
|
|
chr3_-_126646303
|
4.683
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr14_+_19103655
|
4.628
|
NM_023526
|
Nkiras1
|
NFKB inhibitor interacting Ras-like protein 1
|
|
chr7_+_127821093
|
4.581
|
NM_001164580
|
BC030336
|
cDNA sequence BC030336
|
|
chr4_-_6917628
|
4.496
|
NM_145711
|
Tox
|
thymocyte selection-associated high mobility group box
|
|
chr17_-_24826873
|
4.371
|
NM_011522
|
Syngr3
|
synaptogyrin 3
|
|
chr10_-_10802102
|
4.359
|
NM_001114333
NM_016976
|
Grm1
|
glutamate receptor, metabotropic 1
|
|
chr7_+_98239230
|
4.266
|
NM_011807
|
Dlg2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_-_186823350
|
4.203
|
NM_145515
|
Mark1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr18_+_37170365
|
4.080
|
NM_009960
|
Pcdha11
|
protocadherin alpha 11
|
|
chr12_+_30213224
|
4.025
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr1_-_33964439
|
4.019
|
NM_177235
|
Bend6
|
BEN domain containing 6
|
|
chrX_+_140100287
|
3.854
|
NM_001195049
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr1_-_135975681
|
3.846
|
NM_007570
|
Btg2
|
B-cell translocation gene 2, anti-proliferative
|
|
chr6_-_58857000
|
3.819
|
NM_021432
|
Nap1l5
|
nucleosome assembly protein 1-like 5
|
|
chr5_+_75971011
|
3.752
|
NM_001122733
NM_021099
|
Kit
|
kit oncogene
|
|
chr12_+_88019649
|
3.713
|
NM_177354
|
Vash1
|
vasohibin 1
|
|
chr9_+_74823917
|
3.680
|
NM_153584
|
BC031353
|
cDNA sequence BC031353
|
|
chrX_+_139952975
|
3.581
|
NM_001195048
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr10_-_108447674
|
3.498
|
NM_009306
|
Syt1
|
synaptotagmin I
|
|
chr14_-_124591742
|
3.497
|
NM_010201
|
Fgf14
|
fibroblast growth factor 14
|
|
chr12_+_30219769
|
3.484
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr15_-_103170514
|
3.458
|
NM_013866
|
Zfp385a
|
zinc finger protein 385A
|
|
chr9_+_74800828
|
3.453
|
NM_001113283
|
BC031353
|
cDNA sequence BC031353
|
|
chr6_+_134870418
|
3.449
|
NM_009875
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B
|
|
chr4_+_32702401
|
3.397
|
NM_001122978
NM_011997
|
Casp8ap2
|
caspase 8 associated protein 2
|
|
chr13_+_45485106
|
3.395
|
NM_153789
NM_181043
|
Mylip
|
myosin regulatory light chain interacting protein
|
|
chr19_+_36629128
|
3.390
|
NM_001163471
NM_172637
|
Hectd2
|
HECT domain containing 2
|
|
chr14_+_54980488
|
3.377
|
NM_026936
|
Oxa1l
|
oxidase assembly 1-like
|
|
chr7_+_104728398
|
3.239
|
NM_001081267
|
Rsf1
|
remodeling and spacing factor 1
|
|
chr7_-_139978601
|
3.235
|
NM_175268
|
Fam53b
|
family with sequence similarity 53, member B
|
|
chr2_+_102546576
|
3.231
|
NM_001077515
NM_011393
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chrX_+_38754480
|
3.180
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr7_-_140004777
|
3.113
|
NM_212473
|
Fam53b
|
family with sequence similarity 53, member B
|
|
chr2_-_117168600
|
3.110
|
NM_011246
|
Rasgrp1
|
RAS guanyl releasing protein 1
|
|
chr7_-_125273061
|
3.071
|
NM_019419
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr2_-_9970235
|
3.046
|
NM_027748
|
Taf3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor
|
|
chr8_-_49760037
|
2.988
|
NM_001145937
NM_011857
|
Odz3
|
odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr11_-_98407281
|
2.978
|
NM_011771
|
Ikzf3
|
IKAROS family zinc finger 3
|
|
chr6_-_126595818
|
2.951
|
NM_010595
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1
|
|
chr2_+_75670230
|
2.901
|
NM_172666
|
Agps
|
alkylglycerone phosphate synthase
|
|
chr11_+_72832489
|
2.876
|
NM_018883
|
Camkk1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha
|
|
chr2_+_65683823
|
2.784
|
NM_153409
NM_178634
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3
|
|
chr3_-_126701367
|
2.770
|
NM_178655
|
Ank2
|
ankyrin 2, brain
|
|
chr17_+_50648736
|
2.748
|
NM_013880
|
Plcl2
|
phospholipase C-like 2
|
|
chr19_-_28057437
|
2.729
|
NM_001166414
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr2_-_39046224
|
2.705
|
NM_178778
|
Scai
|
suppressor of cancer cell invasion
|
|
chr19_+_60831889
|
2.670
|
NM_178421
|
Nanos1
|
nanos homolog 1 (Drosophila)
|
|
chr18_+_37249789
|
2.669
|
NM_001003671
|
Pcdhac1
|
protocadherin alpha subfamily C, 1
|
|
chr12_-_4234293
|
2.587
|
NM_134046
|
Cenpo
|
centromere protein O
|
|
chr5_+_106305563
|
2.529
|
NM_018759
|
Zfp326
|
zinc finger protein 326
|
|
chr2_+_119722957
|
2.453
|
NM_001164274
NM_013720
|
Mga
|
MAX gene associated
|
|
chr1_-_166388431
|
2.448
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr13_-_54867800
|
2.388
|
NM_033610
|
Sncb
|
synuclein, beta
|
|
chr6_+_86799460
|
2.359
|
NM_001040106
NM_177762
|
Aak1
|
AP2 associated kinase 1
|
|
chr11_+_42233258
|
2.340
|
NM_008070
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2
|
|
chr1_+_194014677
|
2.331
|
NM_001038607
NM_010600
|
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr18_-_16967557
|
2.259
|
NM_007664
|
Cdh2
|
cadherin 2
|
|
chr10_-_59414516
|
2.163
|
NM_029083
|
Ddit4
|
DNA-damage-inducible transcript 4
|
|
chr14_-_125076242
|
2.148
|
NM_207667
|
Fgf14
|
fibroblast growth factor 14
|
|
chr6_+_49269351
|
2.133
|
NM_175098
|
Ccdc126
|
coiled-coil domain containing 126
|
|
chr12_+_4924157
|
2.098
|
NM_001099628
|
Atad2b
|
ATPase family, AAA domain containing 2B
|
|
chr19_-_28085534
|
2.090
|
NM_011265
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr10_-_116018526
|
2.077
|
NM_001037846
NM_001037847
NM_001037848
NM_028082
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr11_-_78510874
|
2.023
|
NM_008702
|
Nlk
|
nemo like kinase
|
|
chr4_+_135728188
|
1.983
|
NM_177733
|
E2f2
|
E2F transcription factor 2
|
|
chr15_-_100416795
|
1.980
|
NM_010127
|
Pou6f1
|
POU domain, class 6, transcription factor 1
|
|
chr10_-_85961630
|
1.980
|
NM_001164495
NM_013722
|
Syn3
|
synapsin III
|
|
chr4_+_34011567
|
1.978
|
NM_007726
|
Cnr1
|
cannabinoid receptor 1 (brain)
|
|
chr1_+_60466462
|
1.967
|
NM_001198570
NM_001198571
NM_198127
|
Abi2
|
abl-interactor 2
|
|
chr5_-_138596915
|
1.960
|
NM_019747
|
Zfp113
|
zinc finger protein 113
|
|
chr18_-_35374663
|
1.958
|
NM_178005
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr11_-_88922061
|
1.913
|
NM_019505
|
Dgke
|
diacylglycerol kinase, epsilon
|
|
chr6_+_107479719
|
1.902
|
NM_008516
|
Lrrn1
|
leucine rich repeat protein 1, neuronal
|
|
chr17_+_24551509
|
1.874
|
NM_001080127
NM_001080128
NM_009070
|
Rnps1
|
ribonucleic acid binding protein S1
|
|
chr9_-_57682040
|
1.865
|
NM_019689
|
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr2_-_37502738
|
1.839
|
NM_009261
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr16_-_91688961
|
1.839
|
NM_021720
|
Donson
|
downstream neighbor of SON
|
|
chr13_-_58229884
|
1.805
|
NM_029872
|
Hnrnpa0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr10_+_28931448
|
1.764
|
NM_001162537
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene
|
|
chr15_-_96472392
|
1.755
|
NM_134086
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr15_-_96473193
|
1.750
|
NM_001166458
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr5_-_121459496
|
1.724
|
NM_011286
|
Rph3a
|
rabphilin 3A
|
|
chr5_+_73882264
|
1.693
|
NM_001190733
NM_178896
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr3_-_62310907
|
1.674
|
NM_028136
|
Dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36
|
|
chr5_+_14025270
|
1.665
|
NM_011348
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr6_-_97129076
|
1.661
|
NM_001081111
|
Tmf1
|
TATA element modulatory factor 1
|
|
chr11_+_96912130
|
1.646
|
NM_001081434
|
Osbpl7
|
oxysterol binding protein-like 7
|
|
chr10_+_79611013
|
1.628
|
NM_021565
|
Midn
|
midnolin
|
|
chr11_-_29925939
|
1.626
|
NM_146016
|
Eml6
|
echinoderm microtubule associated protein like 6
|
|
chr3_+_30691760
|
1.611
|
NM_027016
|
Sec62
|
SEC62 homolog (S. cerevisiae)
|
|
chr2_+_13995715
|
1.596
|
NM_011484
|
Stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1
|
|
chr16_-_10785590
|
1.586
|
NM_009896
|
Socs1
|
suppressor of cytokine signaling 1
|
|
chr5_+_151062363
|
1.567
|
NM_172887
|
Fry
|
furry homolog (Drosophila)
|
|
chr14_+_57506630
|
1.565
|
NM_029498
|
Zmym2
|
zinc finger, MYM-type 2
|
|
chr2_-_25435618
|
1.558
|
NM_001005424
|
Gm996
|
predicted gene 996
|
|
chr5_+_73872293
|
1.555
|
NM_001190734
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr18_+_35758269
|
1.553
|
NM_026420
|
Paip2
|
polyadenylate-binding protein-interacting protein 2
|
|
chr1_+_173697246
|
1.547
|
NM_013730
|
Slamf1
|
signaling lymphocytic activation molecule family member 1
|
|
chr13_-_12199109
|
1.528
|
NM_023868
|
Ryr2
|
ryanodine receptor 2, cardiac
|
|
chr1_-_157820378
|
1.479
|
NM_001039184
|
Cep350
|
centrosomal protein 350
|
|
chr2_+_173563060
|
1.477
|
NM_019806
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C
|
|
chr13_+_120217406
|
1.458
|
NM_001079849
|
Paip1
|
polyadenylate binding protein-interacting protein 1
|
|
chr11_+_102622745
|
1.435
|
NM_001110778
NM_009613
|
Adam11
|
a disintegrin and metallopeptidase domain 11
|
|
chr17_-_47748405
|
1.424
|
NM_016859
|
Bysl
|
bystin-like
|
|
chr3_-_146444377
|
1.399
|
NM_001164200
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr5_+_115461532
|
1.396
|
NM_029012
|
Sppl3
|
signal peptide peptidase 3
|
|
chr1_-_23389950
|
1.371
|
NM_001081079
|
Ogfrl1
|
opioid growth factor receptor-like 1
|
|
chr13_-_21545556
|
1.365
|
NM_001013786
|
Zfp187
|
zinc finger protein 187
|
|
chr4_+_13711928
|
1.358
|
NM_009822
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_-_133843139
|
1.314
|
NM_001163794
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chr11_+_77908168
|
1.300
|
NM_145430
|
BC017647
|
cDNA sequence BC017647
|
|
chr3_+_87329497
|
1.280
|
NM_001083318
NM_012051
|
Etv3
|
ets variant gene 3
|
|
chr3_+_101180976
|
1.279
|
NM_207205
|
Igsf3
|
immunoglobulin superfamily, member 3
|
|
chr9_+_44412931
|
1.278
|
NM_001110826
NM_007841
NM_181324
|
Ddx6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr4_-_148001104
|
1.271
|
NM_001003898
NM_001003899
NM_001008545
NM_001008546
NM_145556
|
Tardbp
|
TAR DNA binding protein
|
|
chr9_+_64949242
|
1.239
|
NM_020043
|
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr6_-_126690651
|
1.218
|
NM_013568
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6
|
|
chr6_-_116143380
|
1.200
|
NM_177412
|
Tmcc1
|
transmembrane and coiled coil domains 1
|
|
chr12_-_92828896
|
1.187
|
NM_175335
|
Gtf2a1
|
general transcription factor II A, 1
|
|
chr10_-_82226774
|
1.170
|
NM_010914
|
Nfyb
|
nuclear transcription factor-Y beta
|
|
chr4_-_133843746
|
1.169
|
NM_146156
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chr17_+_49071906
|
1.156
|
NM_027452
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr3_+_94217197
|
1.135
|
NM_028307
|
Tdrkh
|
tudor and KH domain containing protein
|
|
chr7_+_135888383
|
1.099
|
NM_001029982
|
Sec23ip
|
Sec23 interacting protein
|
|
chr14_-_34260323
|
1.068
|
NM_016700
|
Mapk8
|
mitogen-activated protein kinase 8
|
|
chr14_+_64055230
|
1.053
|
NM_177628
|
Fam167a
|
family with sequence similarity 167, member A
|
|
chr6_-_99616745
|
1.050
|
NM_025829
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3
|
|
chr11_-_103128675
|
1.043
|
NM_016896
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr1_-_38186482
|
1.038
|
NM_019570
|
Rev1
|
REV1 homolog (S. cerevisiae)
|
|
chr4_+_11083585
|
1.013
|
NM_021897
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1
|
|
chr2_+_124436031
|
0.996
|
NM_172537
NM_199238
NM_199239
NM_199240
NM_199241
|
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr5_-_103640272
|
0.974
|
NM_001081567
NM_009158
|
Mapk10
|
mitogen-activated protein kinase 10
|
|
chr15_+_92227240
|
0.964
|
NM_001164593
|
Pdzrn4
|
PDZ domain containing RING finger 4
|
|
chr8_-_48374755
|
0.960
|
NM_001114311
|
Stox2
|
storkhead box 2
|
|
chr10_-_116813919
|
0.939
|
NM_001013391
|
Cpsf6
|
cleavage and polyadenylation specific factor 6
|
|
chr10_-_33344405
|
0.921
|
NM_175448
|
Clvs2
|
clavesin 2
|
|
chr15_-_96472600
|
0.920
|
NM_001166456
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr10_+_40069113
|
0.904
|
NM_001168304
NM_198164
|
Cdk19
|
cyclin-dependent kinase 19
|
|
chr12_-_32398730
|
0.902
|
NM_175191
|
Gpr22
|
G protein-coupled receptor 22
|
|
chr8_-_48437701
|
0.901
|
NM_175162
|
Stox2
|
storkhead box 2
|
|
chr7_-_134757946
|
0.895
|
NM_177226
|
Zfp629
|
zinc finger protein 629
|
|
chr4_+_49072327
|
0.878
|
NM_178756
|
E130309F12Rik
|
RIKEN cDNA E130309F12 gene
|
|
chrX_-_153853209
|
0.857
|
NM_011077
|
Phex
|
phosphate regulating gene with homologies to endopeptidases on the X chromosome (hypophosphatemia, vitamin D resistant rickets)
|
|
chr18_+_37164856
|
0.835
|
NM_009961
|
Pcdha10
|
protocadherin alpha 10
|
|
chr9_-_44607274
|
0.833
|
NM_027865
|
Tmem25
|
transmembrane protein 25
|
|
chr4_+_59818521
|
0.825
|
NM_172468
|
Snx30
|
sorting nexin family member 30
|
|
chrX_-_110299092
|
0.812
|
NM_018818
|
Chm
|
choroidermia
|
|
chr14_-_96918064
|
0.808
|
NM_053105
|
Klhl1
|
kelch-like 1 (Drosophila)
|
|
chr4_+_13670425
|
0.799
|
NM_001111026
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_58810899
|
0.788
|
NM_177855
|
Med12l
|
mediator of RNA polymerase II transcription, subunit 12 homolog (yeast)-like
|
|
chrX_-_159267249
|
0.784
|
NM_178935
|
Txlng
|
taxilin gamma
|
|
chr4_-_98484163
|
0.780
|
NM_172872
|
Kank4
|
KN motif and ankyrin repeat domains 4
|
|
chr5_+_87233514
|
0.767
|
NM_177680
|
Ythdc1
|
YTH domain containing 1
|
|
chr11_-_55125758
|
0.758
|
NM_001029988
|
Fat2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr10_-_122513447
|
0.739
|
NM_001163024
NM_001163025
NM_153395
|
Mon2
|
MON2 homolog (yeast)
|
|
chrX_-_101396356
|
0.734
|
NM_001077354
|
C77370
|
expressed sequence C77370
|
|
chr2_-_37278415
|
0.729
|
NM_001100591
|
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2
|
|
chr10_+_20032269
|
0.712
|
NM_001025392
NM_001025393
NM_153787
|
Bclaf1
|
BCL2-associated transcription factor 1
|
|
chr17_-_45686604
|
0.708
|
NM_001013749
|
Tmem151b
|
transmembrane protein 151B
|