|
chr2_+_136539143
|
63.364
|
NM_011428
|
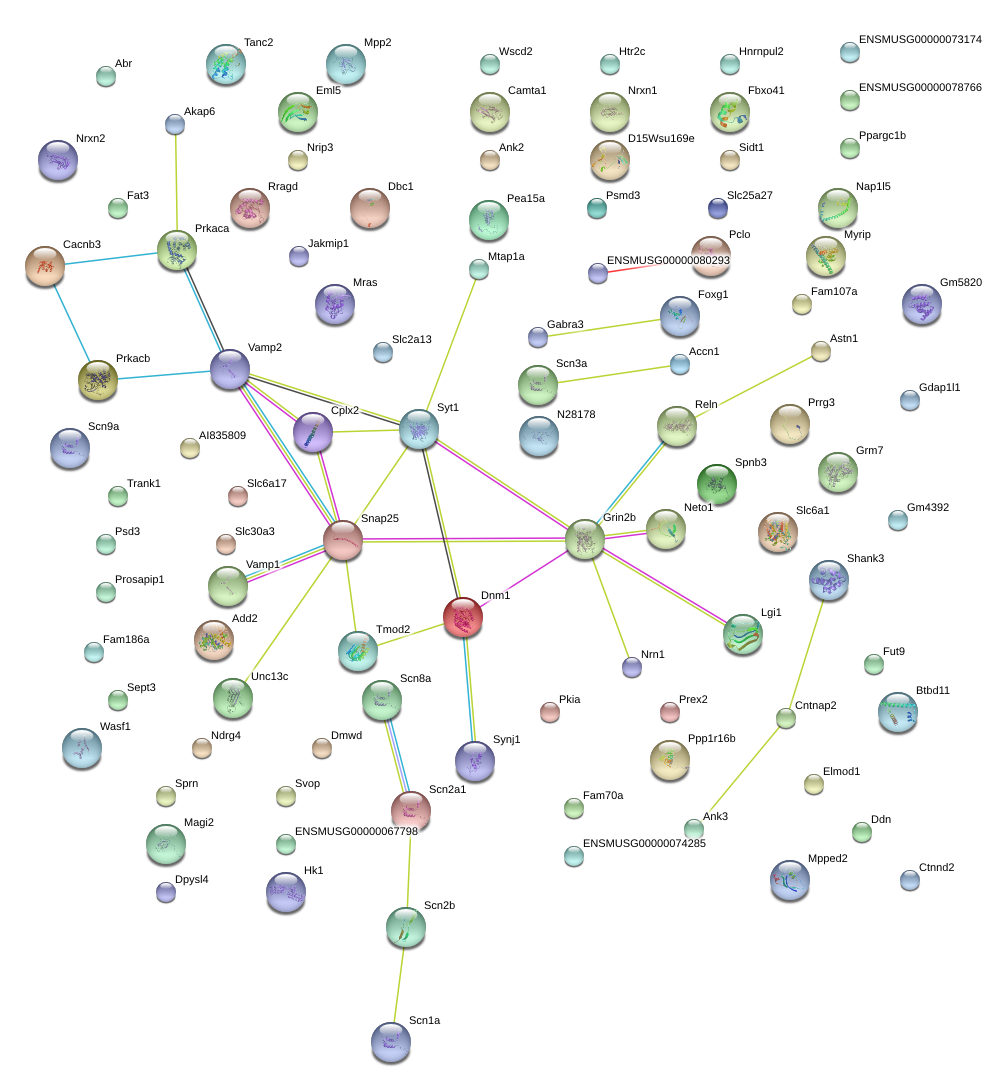
Snap25
|
synaptosomal-associated protein 25
|
|
chr10_-_108447674
|
51.175
|
NM_009306
|
Syt1
|
synaptotagmin I
|
|
chr9_-_53823107
|
47.095
|
NM_177769
|
Elmod1
|
ELMO domain containing 1
|
|
chr15_-_91403685
|
43.718
|
NM_001033633
|
Slc2a13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr15_+_82105322
|
41.228
|
NM_011889
|
Sept3
|
septin 3
|
|
chr10_+_40603330
|
39.062
|
NM_031877
|
Wasf1
|
WASP family 1
|
|
chr7_-_116925058
|
38.857
|
NM_020610
|
Nrip3
|
nuclear receptor interacting protein 3
|
|
chr4_-_49610586
|
38.613
|
NM_025944
|
2810432L12Rik
|
RIKEN cDNA 2810432L12 gene
|
|
chr8_+_98226914
|
36.864
|
NM_001195006
NM_145602
|
Ndrg4
|
N-myc downstream regulated gene 4
|
|
chr13_+_54472705
|
35.544
|
NM_009946
|
Cplx2
|
complexin 2
|
|
chr2_-_130468507
|
33.491
|
NM_197945
|
Prosapip1
|
ProSAPiP1 protein
|
|
chr4_-_25727109
|
33.306
|
NM_010243
|
Fut9
|
fucosyltransferase 9
|
|
chr15_+_30102306
|
33.277
|
NM_008729
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2
|
|
chr2_-_32208737
|
32.896
|
NM_010065
|
Dnm1
|
dynamin 1
|
|
chr11_+_68902025
|
32.595
|
NM_009497
|
Vamp2
|
vesicle-associated membrane protein 2
|
|
chr6_-_58857000
|
31.831
|
NM_021432
|
Nap1l5
|
nucleosome assembly protein 1-like 5
|
|
chr9_+_111214242
|
30.973
|
NM_001164659
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr5_-_114541313
|
30.889
|
NM_026805
|
Svop
|
SV2 related protein
|
|
chr19_+_38339167
|
30.863
|
NM_020278
|
Lgi1
|
leucine-rich repeat LGI family, member 1
|
|
chr3_+_7366586
|
29.941
|
NM_008862
|
Pkia
|
protein kinase inhibitor, alpha
|
|
chr18_+_86564229
|
29.929
|
NM_144946
|
Neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr6_+_47194393
|
29.807
|
NM_025771
|
Cntnap2
|
contactin associated protein-like 2
|
|
chr15_+_89330287
|
29.111
|
NM_021423
|
Shank3
|
SH3/ankyrin domain gene 3
|
|
chr6_+_45010040
|
28.375
|
NM_001004357
|
Cntnap2
|
contactin associated protein-like 2
|
|
chr10_+_69388242
|
28.179
|
NM_009670
NM_170687
|
Ank3
|
ankyrin 3, epithelial
|
|
chr6_+_110595586
|
27.547
|
NM_177328
|
Grm7
|
glutamate receptor, metabotropic 7
|
|
chr13_-_36826245
|
26.078
|
NM_153529
|
Nrn1
|
neuritin 1
|
|
chr10_+_68996455
|
26.036
|
NM_146005
NM_170688
NM_170689
NM_170690
NM_170728
NM_170729
NM_170730
|
Ank3
|
ankyrin 3, epithelial
|
|
chr9_+_44925922
|
25.965
|
NM_001014761
|
Scn2b
|
sodium channel, voltage-gated, type II, beta
|
|
chrX_+_143397055
|
25.519
|
NM_008312
|
Htr2c
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr9_-_73781373
|
24.609
|
NM_001081153
|
Unc13c
|
unc-13 homolog C (C. elegans)
|
|
chr12_+_53800369
|
24.413
|
NM_198111
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr4_-_68615382
|
23.855
|
NM_019967
|
Dbc1
|
deleted in bladder cancer 1 (human)
|
|
chr4_-_151235861
|
23.807
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr1_-_174136874
|
23.757
|
NM_011063
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A
|
|
chr15_+_98462650
|
23.535
|
NM_001044741
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr14_-_9142289
|
23.321
|
NM_183187
|
Fam107a
|
family with sequence similarity 107, member A
|
|
chr5_+_14514906
|
23.283
|
NM_001110796
NM_011995
|
Pclo
|
piccolo (presynaptic cytomatrix protein)
|
|
chr2_+_158492468
|
23.119
|
NM_153089
|
Ppp1r16b
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr11_-_76322920
|
23.044
|
NM_198018
|
Abr
|
active BCR-related gene
|
|
chr7_+_146271899
|
23.023
|
NM_011993
|
Dpysl4
|
dihydropyrimidinase-like 4
|
|
chr15_+_98465193
|
23.017
|
NM_007581
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr3_-_107320881
|
22.785
|
NM_172271
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17
|
|
chr5_+_37442095
|
22.782
|
NM_178394
|
Jakmip1
|
janus kinase and microtubule interacting protein 1
|
|
chr10_+_85061155
|
22.688
|
NM_001017525
|
Btbd11
|
BTB (POZ) domain containing 11
|
|
chr19_+_4711220
|
22.579
|
NM_021287
|
Spnb3
|
spectrin beta 3
|
|
chr15_-_98638348
|
22.415
|
NM_001013741
|
Ddn
|
dendrin
|
|
chr2_+_163264188
|
22.129
|
NM_144891
|
Gdap1l1
|
ganglioside-induced differentiation-associated protein 1-like 1
|
|
chr2_-_66278870
|
22.066
|
NM_018733
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha
|
|
chr5_-_21850490
|
21.937
|
NM_011261
|
Reln
|
reelin
|
|
chr3_-_146444377
|
21.644
|
NM_001164200
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr17_-_43803963
|
21.051
|
NM_028711
|
Slc25a27
|
solute carrier family 25, member 27
|
|
chr5_+_113919352
|
21.012
|
NM_177292
|
Wscd2
|
WSC domain containing 2
|
|
chr6_+_114232618
|
20.581
|
NM_178703
|
Slc6a1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr1_+_160292470
|
20.370
|
NM_007495
|
Astn1
|
astrotactin 1
|
|
chr11_-_76391228
|
19.947
|
NM_198894
|
Abr
|
active BCR-related gene
|
|
chr9_-_16182674
|
19.799
|
NM_001080814
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr19_+_6427922
|
19.687
|
NM_020253
|
Nrxn2
|
neurexin II
|
|
chr12_+_50483869
|
19.508
|
NM_001160112
NM_008241
|
Foxg1
|
forkhead box G1
|
|
chr6_+_86028044
|
19.428
|
NM_013458
|
Add2
|
adducin 2 (beta)
|
|
chr3_-_126646303
|
19.033
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr1_+_11254043
|
18.874
|
NM_001033636
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr10_+_84849558
|
18.814
|
NM_028709
|
Btbd11
|
BTB (POZ) domain containing 11
|
|
chr2_+_121121189
|
18.752
|
NM_001173506
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr9_+_120213190
|
18.579
|
NM_144557
|
Myrip
|
myosin VIIA and Rab interacting protein
|
|
chr2_+_106535757
|
18.571
|
NM_029837
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr11_-_101949793
|
18.466
|
NM_016695
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chrX_+_69208336
|
18.445
|
NM_001081135
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr11_-_80966253
|
18.105
|
NM_007384
|
Accn1
|
amiloride-sensitive cation channel 1, neuronal (degenerin)
|
|
chr2_+_65508501
|
18.044
|
NM_001099298
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1
|
|
chr4_+_33069972
|
17.617
|
NM_027491
|
Rragd
|
Ras-related GTP binding D
|
|
chr2_+_121115337
|
17.544
|
NM_032393
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr16_-_91011339
|
17.445
|
NM_001045515
NM_001164483
|
Synj1
|
synaptojanin 1
|
|
chr16_-_44332950
|
17.399
|
NM_001159419
NM_198034
|
Sidt1
|
SID1 transmembrane family, member 1
|
|
chr7_+_19661548
|
17.377
|
NM_010058
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif
|
|
chr3_-_146433544
|
17.089
|
NM_001164199
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr10_-_61842599
|
16.573
|
NM_010438
|
Hk1
|
hexokinase 1
|
|
chr9_-_75446888
|
16.288
|
NM_001038710
|
Tmod2
|
tropomodulin 2
|
|
chrX_-_35605626
|
16.214
|
NM_172930
|
Fam70a
|
family with sequence similarity 70, member A
|
|
chr12_-_100139666
|
16.028
|
NM_001081191
|
Eml5
|
echinoderm microtubule associated protein like 5
|
|
chr9_-_99336923
|
15.996
|
NM_008624
|
Mras
|
muscle and microspikes RAS
|
|
chr11_+_105451296
|
15.740
|
NM_181071
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr15_-_76648532
|
15.729
|
NM_001168288
NM_198420
|
Arhgap39
|
Rho GTPase activating protein 39
|
|
chrX_-_69901382
|
15.668
|
NM_008067
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3
|
|
chr8_-_70342105
|
15.534
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr6_-_85452851
|
15.384
|
NM_001001160
|
Fbxo41
|
F-box protein 41
|
|
chr4_+_42930076
|
15.276
|
NM_172690
|
N28178
|
expressed sequence N28178
|
|
chr2_+_106533615
|
15.216
|
NM_001143683
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr11_-_81781897
|
15.191
|
NM_001034013
|
Accn1
|
amiloride-sensitive cation channel 1, neuronal (degenerin)
|
|
chr18_-_61560053
|
14.900
|
NM_133249
|
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta
|
|
chr5_+_19413335
|
14.610
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr5_-_31395853
|
14.602
|
NM_011773
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr18_+_39152776
|
14.563
|
NM_175164
|
Arhgap26
|
Rho GTPase activating protein 26
|
|
chr6_-_136123521
|
14.506
|
NM_008171
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2)
|
|
chr7_-_147340532
|
14.374
|
NM_183147
|
Sprn
|
shadow of prion protein
|
|
chr1_-_77511653
|
14.216
|
NM_007936
|
Epha4
|
Eph receptor A4
|
|
chr11_+_75007475
|
14.137
|
NM_177708
|
Rtn4rl1
|
reticulon 4 receptor-like 1
|
|
chr10_-_61802889
|
14.005
|
NM_001146100
|
Hk1
|
hexokinase 1
|
|
chr2_+_38206759
|
13.958
|
NM_010710
|
Lhx2
|
LIM homeobox protein 2
|
|
chr3_-_126701367
|
13.893
|
NM_178655
|
Ank2
|
ankyrin 2, brain
|
|
chr16_-_34514112
|
13.841
|
NM_001164268
NM_177357
|
Kalrn
|
kalirin, RhoGEF kinase
|
|
chr2_+_29658139
|
13.799
|
NM_011989
|
Slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr3_-_146433200
|
13.515
|
NM_001164198
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr14_-_66043991
|
13.470
|
NM_010932
|
Pnoc
|
prepronociceptin
|
|
chr3_+_105255247
|
13.441
|
NM_001039347
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3
|
|
chr9_-_75459125
|
13.437
|
NM_016711
|
Tmod2
|
tropomodulin 2
|
|
chr7_-_133942605
|
13.343
|
NM_001177307
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr5_+_18732863
|
13.282
|
NM_001170746
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr11_-_76384852
|
13.114
|
NM_198895
|
Abr
|
active BCR-related gene
|
|
chr2_+_20431673
|
13.114
|
NM_029895
NM_178059
|
Etl4
|
enhancer trap locus 4
|
|
chr1_-_137481914
|
13.065
|
NM_173437
|
Nav1
|
neuron navigator 1
|
|
chr2_+_20211539
|
13.044
|
NM_001081006
|
Etl4
|
enhancer trap locus 4
|
|
chr12_-_71448600
|
13.028
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr1_-_37776633
|
13.028
|
NM_028096
|
2010300C02Rik
|
RIKEN cDNA 2010300C02 gene
|
|
chr2_-_12932442
|
13.015
|
NM_153155
|
C1ql3
|
C1q-like 3
|
|
chr6_-_126595818
|
12.954
|
NM_010595
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1
|
|
chr6_+_103460834
|
12.921
|
NM_007697
|
Chl1
|
cell adhesion molecule with homology to L1CAM
|
|
chr12_-_109241423
|
12.588
|
NM_001079883
NM_021399
|
Bcl11b
|
B-cell leukemia/lymphoma 11B
|
|
chr11_-_97914582
|
12.484
|
NM_146028
|
Stac2
|
SH3 and cysteine rich domain 2
|
|
chr7_-_133942767
|
12.470
|
NM_007438
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr11_+_116780161
|
11.963
|
NM_172948
|
Mgat5b
|
mannoside acetylglucosaminyltransferase 5, isoenzyme B
|
|
chr12_-_32398730
|
11.909
|
NM_175191
|
Gpr22
|
G protein-coupled receptor 22
|
|
chr1_-_134439500
|
11.907
|
NM_177129
|
Cntn2
|
contactin 2
|
|
chr9_-_50535949
|
11.524
|
NM_178118
|
Dixdc1
|
DIX domain containing 1
|
|
chr5_+_34338625
|
11.449
|
NM_001001985
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr11_+_62094922
|
11.332
|
NM_028360
NM_029704
|
Ttc19
|
tetratricopeptide repeat domain 19
|
|
chr6_-_122692754
|
11.228
|
NM_011401
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr11_-_80191504
|
11.215
|
NM_027472
|
5730455P16Rik
|
RIKEN cDNA 5730455P16 gene
|
|
chr6_-_82889694
|
11.119
|
NM_001113481
NM_011350
|
Sema4f
|
sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain
|
|
chr1_+_75396650
|
11.117
|
NM_001173477
|
Speg
|
SPEG complex locus
|
|
chr6_+_115084919
|
10.970
|
NM_001111015
NM_013681
|
Syn2
|
synapsin II
|
|
chr1_-_58642994
|
10.773
|
NM_172513
|
Fam126b
|
family with sequence similarity 126, member B
|
|
chr3_+_94282752
|
10.741
|
NM_172434
|
Celf3
|
VCUGBP, Elav-like family member 3
|
|
chr7_+_28043740
|
10.348
|
NM_010950
|
Numbl
|
numb-like
|
|
chr10_+_4424140
|
10.346
|
NM_001161822
NM_019958
|
Rgs17
|
regulator of G-protein signaling 17
|
|
chr2_-_91803660
|
10.341
|
NM_138306
|
Dgkz
|
diacylglycerol kinase zeta
|
|
chr3_-_138738152
|
10.247
|
NM_001040690
NM_145544
|
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1
|
|
chr7_+_149135647
|
10.203
|
NM_029426
|
Brsk2
|
BR serine/threonine kinase 2
|
|
chr17_-_37088578
|
10.202
|
NM_029632
|
Ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11
|
|
chr7_-_133943848
|
10.128
|
NM_001177308
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr1_-_39707919
|
9.847
|
NM_170597
|
Creg2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr2_+_91490266
|
9.803
|
NM_001145902
|
Arhgap1
|
Rho GTPase activating protein 1
|
|
chr2_-_29979973
|
9.731
|
NM_178694
|
Zer1
|
zer-1 homolog (C. elegans)
|
|
chr1_-_159342713
|
9.668
|
NM_177644
|
Rasal2
|
RAS protein activator like 2
|
|
chr2_-_91790504
|
9.563
|
NM_001166597
|
Dgkz
|
diacylglycerol kinase zeta
|
|
chr10_+_94039007
|
9.425
|
NM_177026
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr2_+_158492854
|
9.421
|
NM_001159662
|
Ppp1r16b
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr10_-_109437274
|
9.412
|
NM_001081035
|
Nav3
|
neuron navigator 3
|
|
chr5_-_107125783
|
9.348
|
NM_026856
|
Zfp644
|
zinc finger protein 644
|
|
chr8_+_125935441
|
9.331
|
NM_023279
|
Tubb3
|
tubulin, beta 3
|
|
chr11_-_69373707
|
9.275
|
NM_007911
|
Efnb3
|
ephrin B3
|
|
chr17_-_51965912
|
9.257
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_67703726
|
9.205
|
NM_008984
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M
|
|
chr8_-_70434808
|
9.201
|
NM_030263
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr2_+_58998885
|
8.734
|
NM_026361
NM_175464
|
Pkp4
|
plakophilin 4
|
|
chr12_-_51750204
|
8.699
|
NM_008858
|
Prkd1
|
protein kinase D1
|
|
chr11_-_119409041
|
8.618
|
NM_008730
|
Nptx1
|
neuronal pentraxin 1
|
|
chr8_+_119467267
|
8.606
|
NM_177700
|
Atmin
|
ATM interactor
|
|
chr3_+_27081918
|
8.555
|
NM_178772
|
Nceh1
|
arylacetamide deacetylase-like 1
|
|
chr11_+_104092749
|
8.384
|
NM_001038609
NM_010838
|
Mapt
|
microtubule-associated protein tau
|
|
chr11_-_119903061
|
8.374
|
NM_001198787
|
Aatk
|
apoptosis-associated tyrosine kinase
|
|
chr2_+_164793487
|
8.370
|
NM_020333
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr6_-_13789847
|
8.273
|
NM_145066
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr10_+_94038001
|
8.222
|
NM_001168684
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr16_-_52453104
|
8.161
|
NM_009655
|
Alcam
|
activated leukocyte cell adhesion molecule
|
|
chr6_+_29718424
|
8.078
|
NM_001171000
NM_021414
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2
|
|
chr14_-_47441795
|
8.051
|
NM_022023
|
Gmfb
|
glia maturation factor, beta
|
|
chr10_-_80945445
|
8.030
|
NM_176954
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr11_-_74710562
|
8.007
|
NM_197943
|
Sgsm2
|
small G protein signaling modulator 2
|
|
chr7_+_98239230
|
7.893
|
NM_011807
|
Dlg2
|
discs, large homolog 2 (Drosophila)
|
|
chr10_+_4795808
|
7.742
|
NM_153399
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr15_+_16707855
|
7.719
|
NM_009869
|
Cdh9
|
cadherin 9
|
|
chr19_+_6046575
|
7.631
|
NM_028769
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin
|
|
chr7_+_64845887
|
7.623
|
NM_001038701
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3
|
|
chr8_+_32200076
|
7.606
|
NM_025869
|
Dusp26
|
dual specificity phosphatase 26 (putative)
|
|
chr18_-_31606908
|
7.541
|
NM_009308
|
Syt4
|
synaptotagmin IV
|
|
chr13_+_58907953
|
7.510
|
NM_008745
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr18_-_35374663
|
7.438
|
NM_178005
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr17_-_82137708
|
7.260
|
NM_001112798
NM_011406
|
Slc8a1
D830013E24Rik
|
solute carrier family 8 (sodium/calcium exchanger), member 1
RIKEN cDNA D830013E24 gene
|
|
chr9_-_64101354
|
7.221
|
NM_008927
|
Map2k1
|
mitogen-activated protein kinase kinase 1
|
|
chr4_-_45097427
|
7.139
|
NM_001024142
|
Fbxo10
|
F-box protein 10
|
|
chr1_+_37356722
|
7.086
|
NM_030266
|
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I
|
|
chr2_-_117168600
|
6.882
|
NM_011246
|
Rasgrp1
|
RAS guanyl releasing protein 1
|
|
chr19_-_28057437
|
6.863
|
NM_001166414
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr7_-_148615247
|
6.833
|
NM_021316
|
Cend1
|
cell cycle exit and neuronal differentiation 1
|
|
chrX_-_133403050
|
6.782
|
NM_176971
|
Rab9b
|
RAB9B, member RAS oncogene family
|
|
chr8_-_114457667
|
6.725
|
NM_001033321
|
Tmem231
|
transmembrane protein 231
|
|
chr9_+_26559146
|
6.640
|
NM_029792
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr5_-_72049479
|
6.633
|
NM_010251
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4
|
|
chr7_+_133415982
|
6.621
|
NM_145587
|
Sbk1
|
SH3-binding kinase 1
|
|
chr11_-_119908236
|
6.524
|
NM_001198785
NM_007377
|
Aatk
|
apoptosis-associated tyrosine kinase
|
|
chr18_+_65739883
|
6.428
|
NM_207255
|
Zfp532
|
zinc finger protein 532
|
|
chr8_-_72963464
|
6.426
|
NM_001004062
|
Crtc1
|
CREB regulated transcription coactivator 1
|
|
chr1_+_10983542
|
6.304
|
NM_029525
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr15_-_66801198
|
6.188
|
NM_008681
|
Ndrg1
|
N-myc downstream regulated gene 1
|
|
chr7_+_146132386
|
6.183
|
NM_028708
|
Jakmip3
|
janus kinase and microtubule interacting protein 3
|
|
chr2_+_35477498
|
6.153
|
NM_001114124
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr5_+_37259780
|
6.148
|
NM_172994
|
Ppp2r2c
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), gamma isoform
|
|
chr12_+_25659777
|
5.978
|
NM_001081378
|
Kidins220
|
kinase D-interacting substrate 220
|
|
chr11_-_96691261
|
5.917
|
NM_008686
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1
|