|
chr1_+_138027943
|
15.652
|
NM_001039472
|
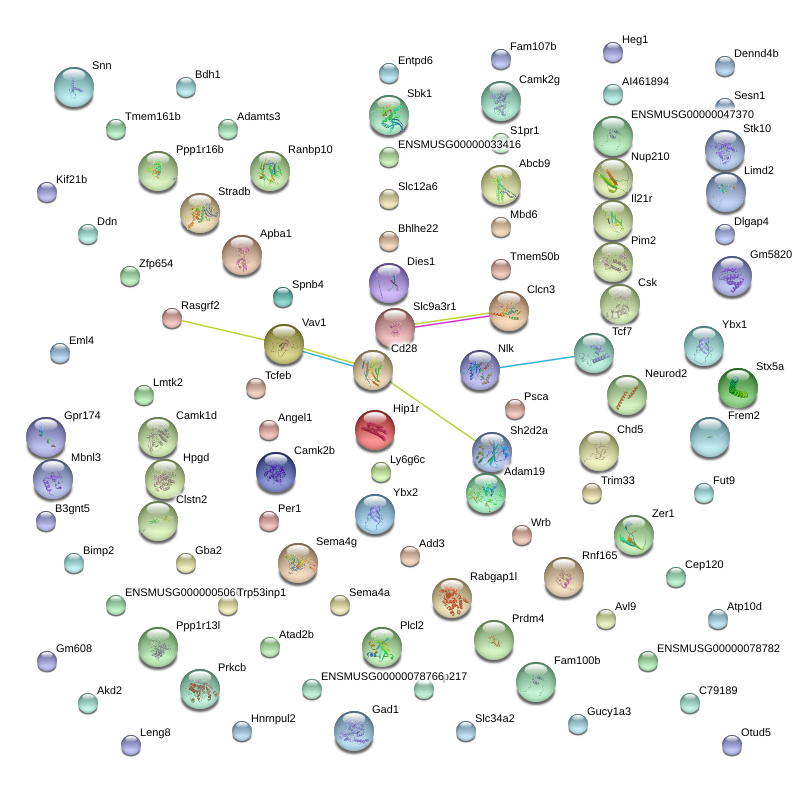
Kif21b
|
kinesin family member 21B
|
|
chr3_-_88265103
|
13.931
|
NM_001163489
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr3_-_88262805
|
13.914
|
NM_001163491
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr7_+_129432585
|
13.903
|
NM_008855
|
Prkcb
|
protein kinase C, beta
|
|
chr3_-_88259682
|
13.332
|
NM_013658
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr3_-_88262982
|
13.209
|
NM_001163490
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr4_-_43591721
|
11.880
|
NM_172692
|
Gba2
|
glucosidase beta 2
|
|
chr1_-_162281701
|
11.711
|
NM_001038621
|
Rabgap1l
|
RAB GTPase activating protein 1-like
|
|
chr2_+_158492468
|
10.142
|
NM_153089
|
Ppp1r16b
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr2_-_169956719
|
10.097
|
NM_001033299
|
Zfp217
|
zinc finger protein 217
|
|
chr11_-_52095745
|
9.764
|
NM_009331
|
Tcf7
|
transcription factor 7, T-cell specific
|
|
chr6_-_91066809
|
9.490
|
NM_018815
|
Nup210
|
nucleoporin 210
|
|
chr2_-_169968174
|
9.072
|
NM_001159683
|
Zfp217
|
zinc finger protein 217
|
|
chr16_+_44173356
|
8.380
|
NM_001029889
|
Gm608
|
predicted gene 608
|
|
chrX_+_104451216
|
8.348
|
NM_001177781
NM_001033251
|
Gpr174
|
G protein-coupled receptor 174
|
|
chr3_+_103083215
|
7.500
|
NM_001079830
NM_053170
|
Trim33
|
tripartite motif-containing 33
|
|
chrX_+_104479684
|
7.489
|
NM_001177782
|
Gpr174
|
G protein-coupled receptor 174
|
|
chr11_+_69749297
|
7.201
|
NM_016875
|
Ybx2
|
Y box protein 2
|
|
chr16_+_19760300
|
7.197
|
NM_001159407
NM_001159408
NM_054052
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr15_-_98638348
|
7.187
|
NM_001013741
|
Ddn
|
dendrin
|
|
chr15_+_74545268
|
7.066
|
NM_028216
|
Psca
|
prostate stem cell antigen
|
|
chr1_+_59030365
|
6.680
|
NM_172656
|
Stradb
|
STE20-related kinase adaptor beta
|
|
chr11_+_68912444
|
6.675
|
NM_011065
|
Per1
|
period homolog 1 (Drosophila)
|
|
chr3_+_87650676
|
6.578
|
NM_001025571
NM_021309
|
Sh2d2a
|
SH2 domain protein 2A
|
|
chr19_+_23833310
|
6.461
|
NM_177034
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1
|
|
chr10_+_59809598
|
6.337
|
NM_001159572
NM_028732
|
4632428N05Rik
|
RIKEN cDNA 4632428N05 gene
|
|
chr3_+_17954324
|
6.179
|
NM_021560
|
Bhlhe22
|
basic helix-loop-helix family, member e22
|
|
chr2_-_29979973
|
6.133
|
NM_178694
|
Zer1
|
zer-1 homolog (C. elegans)
|
|
chr5_+_53440591
|
6.070
|
NM_011402
|
Slc34a2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr11_-_98190774
|
5.980
|
NM_010895
|
Neurod2
|
neurogenic differentiation 2
|
|
chr11_+_68913653
|
5.945
|
NM_001159367
|
Per1
|
period homolog 1 (Drosophila)
|
|
chr2_+_70400179
|
5.885
|
NM_008077
|
Gad1
|
glutamic acid decarboxylase 1
|
|
chr19_+_8815893
|
5.700
|
NM_001167799
|
Stx5a
|
syntaxin 5A
|
|
chr2_+_150574777
|
5.665
|
NM_172117
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6
|
|
chr7_+_133415982
|
5.605
|
NM_145587
|
Sbk1
|
SH3-binding kinase 1
|
|
chr2_+_158492854
|
5.589
|
NM_001159662
|
Ppp1r16b
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr1_+_60803231
|
5.488
|
NM_007642
|
Cd28
|
CD28 antigen
|
|
chr10_+_41530379
|
5.400
|
NM_001162908
|
Sesn1
|
sestrin 1
|
|
chr16_+_11066390
|
5.374
|
NM_009223
|
Snn
|
stannin
|
|
chr13_-_92901406
|
5.324
|
NM_009027
|
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2
|
|
chr19_+_53217238
|
5.300
|
NM_001164100
|
Add3
|
adducin 3 (gamma)
|
|
chr19_+_8816292
|
5.153
|
NM_019829
|
Stx5a
|
syntaxin 5A
|
|
chr5_+_72594551
|
5.152
|
NM_153389
|
Atp10d
|
ATPase, class V, type 10D
|
|
chr5_+_144861227
|
4.979
|
NM_001081109
|
Lmtk2
|
lemur tyrosine kinase 2
|
|
chr11_+_115024626
|
4.915
|
NM_012030
|
Slc9a3r1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr17_+_47874876
|
4.828
|
NM_001161723
|
Tcfeb
|
transcription factor EB
|
|
chr10_+_41607235
|
4.795
|
NM_001013370
|
Sesn1
|
sestrin 1
|
|
chr11_-_5965743
|
4.768
|
NM_001174053
NM_001174054
NM_007595
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr7_-_28231535
|
4.711
|
NM_032610
|
Spnb4
|
spectrin beta 4
|
|
chr6_+_72297294
|
4.684
|
NM_026696
|
0610030E20Rik
|
RIKEN cDNA 0610030E20 gene
|
|
chr3_+_90070455
|
4.682
|
NM_201407
|
Dennd4b
|
DENN/MADD domain containing 4B
|
|
chr8_-_63433528
|
4.667
|
NM_173876
|
Clcn3
|
chloride channel 3
|
|
chr7_+_132746942
|
4.663
|
NM_021887
|
Il21r
|
interleukin 21 receptor
|
|
chr16_+_33684551
|
4.622
|
NM_175256
|
Heg1
|
HEG homolog 1 (zebrafish)
|
|
chr17_+_47922668
|
4.598
|
NM_011549
|
Tcfeb
|
transcription factor EB
|
|
chr12_-_88067400
|
4.579
|
NM_144524
|
Angel1
|
angel homolog 1 (Drosophila)
|
|
chr19_+_45063750
|
4.569
|
NM_011976
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr3_-_115417932
|
4.552
|
NM_007901
|
S1pr1
|
sphingosine-1-phosphate receptor 1
|
|
chr11_+_45869473
|
4.549
|
NM_009616
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta)
|
|
chr13_+_84361874
|
4.535
|
NM_175187
|
Tmem161b
|
transmembrane protein 161B
|
|
chr12_+_4924157
|
4.517
|
NM_001099628
|
Atad2b
|
ATPase family, AAA domain containing 2B
|
|
chr1_+_9591347
|
4.490
|
NM_178399
|
3110035E14Rik
|
RIKEN cDNA 3110035E14 gene
|
|
chr6_+_56664888
|
4.428
|
NM_030235
|
Avl9
|
AVL9 homolog (S. cerevisiase)
|
|
chr18_+_39152776
|
4.415
|
NM_175164
|
Arhgap26
|
Rho GTPase activating protein 26
|
|
chr19_+_53218486
|
4.381
|
NM_013758
|
Add3
|
adducin 3 (gamma)
|
|
chr4_+_11083585
|
4.371
|
NM_021897
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1
|
|
chr5_-_124545776
|
4.340
|
NM_019875
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr17_+_50648736
|
4.314
|
NM_013880
|
Plcl2
|
phospholipase C-like 2
|
|
chr17_+_47873941
|
4.307
|
NM_001161722
|
Tcfeb
|
transcription factor EB
|
|
chr11_+_32433265
|
4.235
|
NM_009288
|
Stk10
|
serine/threonine kinase 10
|
|
chr10_-_126725826
|
4.176
|
NM_033072
|
Mbd6
|
methyl-CpG binding domain protein 6
|
|
chr8_-_108351060
|
4.166
|
NM_145824
|
Ranbp10
|
RAN binding protein 10
|
|
chr4_+_151712739
|
4.136
|
NM_001081376
NM_029216
|
Chd5
|
chromodomain helicase DNA binding protein 5
|
|
chrX_+_7455387
|
4.093
|
NM_138606
|
Pim2
|
proviral integration site 2
|
|
chrX_-_48559008
|
4.087
|
NM_134163
|
Mbnl3
|
muscleblind-like 3 (Drosophila)
|
|
chr3_-_53461276
|
4.047
|
NM_172862
|
Frem2
|
Fras1 related extracellular matrix protein 2
|
|
chr18_-_53904193
|
4.022
|
NM_178686
|
Cep120
|
centrosomal protein 120
|
|
chr5_+_124423632
|
3.960
|
NM_145070
|
Hip1r
|
huntingtin interacting protein 1 related
|
|
chr7_+_4088622
|
3.954
|
NM_172736
|
Leng8
|
leukocyte receptor cluster (LRC) member 8
|
|
chr14_+_33669941
|
3.870
|
NM_178791
|
E130203B14Rik
|
RIKEN cDNA E130203B14 gene
|
|
chr11_+_116295406
|
3.837
|
NM_176902
|
Fam100b
|
family with sequence similarity 100, member B
|
|
chr8_-_63462080
|
3.816
|
NM_007711
NM_173873
NM_173874
|
Clcn3
|
chloride channel 3
|
|
chr2_+_3630729
|
3.805
|
NM_025626
|
Fam107b
|
family with sequence similarity 107, member B
|
|
chr17_+_57418517
|
3.791
|
NM_001163815
NM_001163816
NM_011691
|
Vav1
|
vav 1 oncogene
|
|
chr9_-_57492978
|
3.790
|
NM_007783
|
Csk
|
c-src tyrosine kinase
|
|
chr2_+_112106470
|
3.759
|
NM_133649
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr2_-_5635547
|
3.705
|
NM_177343
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr16_-_64786146
|
3.688
|
NM_028059
|
Zfp654
|
zinc finger protein 654
|
|
chr10_-_74980017
|
3.680
|
NM_175133
|
1110038D17Rik
|
RIKEN cDNA 1110038D17 gene
|
|
chrX_+_7418894
|
3.652
|
NM_138604
|
Otud5
|
OTU domain containing 5
|
|
chr4_-_25727109
|
3.651
|
NM_010243
|
Fut9
|
fucosyltransferase 9
|
|
chr17_+_83750246
|
3.623
|
NM_001114361
NM_001114362
NM_199466
|
Eml4
|
echinoderm microtubule associated protein like 4
|
|
chr17_+_35204268
|
3.595
|
NM_023463
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr19_+_53214934
|
3.565
|
NM_001164099
NM_001164101
|
Add3
|
adducin 3 (gamma)
|
|
chr3_-_81949667
|
3.563
|
NM_021896
|
Gucy1a3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr5_-_90312358
|
3.562
|
NM_001081401
NM_177872
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3
|
|
chr2_+_156546694
|
3.557
|
NM_001042488
|
Dlgap4
|
discs, large homolog-associated protein 4 (Drosophila)
|
|
chr11_-_106021399
|
3.552
|
NM_172397
|
Limd2
|
LIM domain containing 2
|
|
chr9_-_97933533
|
3.502
|
NM_022319
|
Clstn2
|
calsyntenin 2
|
|
chr18_-_77803843
|
3.479
|
NM_001164504
|
8030462N17Rik
Rnf165
|
RIKEN cDNA 8030462N17 gene
ring finger protein 165
|
|
chr11_-_78510874
|
3.401
|
NM_008702
|
Nlk
|
nemo like kinase
|
|
chr16_+_31422377
|
3.387
|
NM_175177
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr7_+_19946564
|
3.380
|
NM_001010836
|
Ppp1r13l
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like
|
|
chr16_-_91597886
|
3.348
|
NM_030018
|
Tmem50b
|
transmembrane protein 50B
|
|
chr6_+_55401971
|
3.309
|
NM_001025372
NM_007407
|
Adcyap1r1
|
adenylate cyclase activating polypeptide 1 receptor 1
|
|
chr14_+_84843366
|
3.289
|
NM_001013753
|
Pcdh17
|
protocadherin 17
|
|
chr1_-_138130840
|
3.285
|
NM_028872
|
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene
|
|
chr6_-_126690651
|
3.273
|
NM_013568
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6
|
|
chr11_+_83664370
|
3.270
|
NM_009330
|
Hnf1b
|
HNF1 homeobox B
|
|
chr18_-_38094995
|
3.264
|
NM_007858
|
Diap1
|
diaphanous homolog 1 (Drosophila)
|
|
chr11_-_100217433
|
3.264
|
NM_010404
NM_177981
|
Hap1
|
huntingtin-associated protein 1
|
|
chrX_-_137135022
|
3.263
|
NM_001077364
|
Tsc22d3
|
TSC22 domain family, member 3
|
|
chr7_-_31230577
|
3.240
|
NM_007467
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr3_+_94641479
|
3.203
|
NM_001165948
NM_172683
|
Pogz
|
pogo transposable element with ZNF domain
|
|
chr15_-_98748371
|
3.200
|
NM_029098
|
Lmbr1l
|
limb region 1 like
|
|
chr1_+_132613903
|
3.187
|
NM_178691
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae)
|
|
chr6_-_87761537
|
3.135
|
NM_001039394
|
Rab43
|
RAB43, member RAS oncogene family
|
|
chr11_+_83223573
|
3.126
|
NM_001013386
|
Rasl10b
|
RAS-like, family 10, member B
|
|
chr11_+_54117299
|
3.126
|
NM_001033598
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr1_-_74590801
|
3.094
|
NM_176972
|
Usp37
|
ubiquitin specific peptidase 37
|
|
chr3_+_94758936
|
3.089
|
NM_017395
|
Rfx5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr11_+_87574041
|
3.084
|
NM_172449
|
Bzrap1
|
benzodiazepine receptor associated protein 1
|
|
chr15_-_101755120
|
3.020
|
NM_008475
|
Krt4
|
keratin 4
|
|
chr18_-_35900005
|
3.019
|
NM_028261
|
Tmem173
|
transmembrane protein 173
|
|
chr17_-_25974021
|
3.013
|
NM_144816
|
Rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chrX_-_137077639
|
2.976
|
NM_010286
|
Tsc22d3
|
TSC22 domain family, member 3
|
|
chr11_+_54117703
|
2.957
|
NM_001033597
NM_144823
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr5_+_34064372
|
2.954
|
NM_008010
NM_001163215
|
Fgfr3
|
fibroblast growth factor receptor 3
|
|
chr16_+_31428833
|
2.928
|
NM_001122683
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr2_+_119625249
|
2.914
|
NM_019392
|
Tyro3
|
TYRO3 protein tyrosine kinase 3
|
|
chr14_-_70132309
|
2.897
|
NM_134078
|
Chmp7
|
CHMP family, member 7
|
|
chr8_+_74685098
|
2.894
|
NM_023126
|
Rab8a
|
RAB8A, member RAS oncogene family
|
|
chr5_+_115879617
|
2.893
|
NM_008629
|
Msi1
|
Musashi homolog 1(Drosophila)
|
|
chr4_-_63283448
|
2.890
|
NM_029324
|
1700018C11Rik
|
RIKEN cDNA 1700018C11 gene
|
|
chr11_+_56825192
|
2.850
|
NM_001113325
NM_008165
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1)
|
|
chr15_-_100467247
|
2.843
|
NM_174992
NM_001033872
|
Smagp
|
small cell adhesion glycoprotein
|
|
chrX_+_7215705
|
2.832
|
NM_009305
|
Syp
|
synaptophysin
|
|
chr11_-_102157904
|
2.824
|
NM_001098836
NM_001098837
|
Atxn7l3
|
ataxin 7-like 3
|
|
chr4_-_75779379
|
2.817
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr14_+_56179739
|
2.813
|
NM_001199009
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr18_-_47528483
|
2.798
|
NM_018744
|
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr15_+_96117952
|
2.721
|
NM_175251
|
Arid2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr10_+_5151833
|
2.721
|
NM_001079686
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr10_+_79611013
|
2.717
|
NM_021565
|
Midn
|
midnolin
|
|
chr15_+_99532698
|
2.705
|
NM_031842
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr16_+_36071694
|
2.698
|
NM_001159421
NM_198645
|
Ccdc58
|
coiled-coil domain containing 58
|
|
chr1_-_136131981
|
2.665
|
NM_001008533
NM_001039510
|
Adora1
|
adenosine A1 receptor
|
|
chr7_+_108599740
|
2.645
|
NM_001143849
|
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr2_+_112124767
|
2.642
|
NM_133648
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr11_-_69419419
|
2.637
|
NM_013415
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr14_+_56178853
|
2.622
|
NM_133734
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr18_+_42435006
|
2.606
|
NM_172626
|
Rbm27
|
RNA binding motif protein 27
|
|
chr11_+_106650565
|
2.606
|
NM_001166685
NM_177088
|
Ccdc45
|
coiled-coil domain containing 45
|
|
chr12_-_88729144
|
2.606
|
NM_011479
|
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr7_+_104480819
|
2.602
|
NM_001039039
|
Kctd21
|
potassium channel tetramerisation domain containing 21
|
|
chr7_+_108570211
|
2.592
|
NM_001008548
NM_001143848
|
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr3_-_105604199
|
2.580
|
NM_145541
|
Rap1a
|
RAS-related protein-1a
|
|
chr1_+_163072784
|
2.564
|
NM_028664
|
Ankrd45
|
ankyrin repeat domain 45
|
|
chr6_+_113342490
|
2.549
|
NM_133923
|
Ttll3
|
tubulin tyrosine ligase-like family, member 3
|
|
chr1_+_168184269
|
2.546
|
NM_001164528
|
Ildr2
|
immunoglobulin-like domain containing receptor 2
|
|
chr4_-_132895228
|
2.545
|
NM_199306
|
Wdtc1
|
WD and tetratricopeptide repeats 1
|
|
chr3_-_100293092
|
2.523
|
NM_001142952
|
Fam46c
|
family with sequence similarity 46, member C
|
|
chr2_+_156547014
|
2.515
|
NM_001042487
|
Dlgap4
|
discs, large homolog-associated protein 4 (Drosophila)
|
|
chr17_+_49071906
|
2.508
|
NM_027452
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr2_-_7317824
|
2.490
|
NM_001110231
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr14_-_61656804
|
2.469
|
NM_001164155
|
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chrX_+_38754480
|
2.464
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr7_-_106387077
|
2.464
|
NM_177448
|
Mogat2
|
monoacylglycerol O-acyltransferase 2
|
|
chr18_+_35000283
|
2.463
|
NM_144865
|
Reep2
|
receptor accessory protein 2
|
|
chr7_-_59261060
|
2.443
|
NM_001160345
|
Svip
|
small VCP/p97-interacting protein
|
|
chr2_-_65405546
|
2.428
|
NM_018732
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha
|
|
chr19_+_18824472
|
2.386
|
NM_153417
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr17_-_24346280
|
2.359
|
NM_010947
|
Ntn3
|
netrin 3
|
|
chr15_-_88979923
|
2.347
|
NM_011161
|
Mapk11
|
mitogen-activated protein kinase 11
|
|
chr9_-_121614076
|
2.340
|
NM_001164562
NM_178677
|
Sec22c
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae)
|
|
chr2_-_155518064
|
2.321
|
NM_001163452
NM_019828
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr3_-_141645214
|
2.300
|
NM_007560
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B
|
|
chr11_-_86171026
|
2.283
|
NM_001080931
|
Med13
|
mediator complex subunit 13
|
|
chr16_-_37539763
|
2.281
|
NM_028812
|
Gtf2e1
|
general transcription factor II E, polypeptide 1 (alpha subunit)
|
|
chr7_+_106683983
|
2.279
|
NM_177231
NM_178220
|
Arrb1
|
arrestin, beta 1
|
|
chr18_+_21159829
|
2.255
|
NM_019706
NM_207623
|
Rnf138
|
ring finger protein 138
|
|
chr14_-_61665691
|
2.234
|
NM_013869
|
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr12_-_102146390
|
2.230
|
NM_001177673
NM_175493
|
Gpr68
|
G protein-coupled receptor 68
|
|
chr9_+_30838224
|
2.228
|
NM_001115130
NM_172765
|
Zbtb44
|
zinc finger and BTB domain containing 44
|
|
chr2_+_160471623
|
2.224
|
NM_009408
|
Top1
|
topoisomerase (DNA) I
|
|
chr7_+_56214442
|
2.213
|
NM_001111016
|
Nav2
|
neuron navigator 2
|
|
chr18_-_32109233
|
2.206
|
NM_001025381
|
Gpr17
|
G protein-coupled receptor 17
|
|
chr11_-_94622495
|
2.197
|
NM_001105561
|
Gm11545
|
predicted gene 11545
|
|
chr9_-_21116749
|
2.190
|
NM_001110300
NM_009678
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit
|
|
chr11_+_54128404
|
2.183
|
NM_001033599
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr3_-_108003697
|
2.171
|
NM_001081320
NM_028061
|
Cyb561d1
|
cytochrome b-561 domain containing 1
|
|
chr5_+_137960107
|
2.136
|
NM_031408
|
Gigyf1
|
GRB10 interacting GYF protein 1
|
|
chr7_-_29221173
|
2.110
|
NM_175021
|
Samd4b
|
sterile alpha motif domain containing 4B
|
|
chr7_-_106859849
|
2.110
|
NM_175316
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1
|
|
chr14_+_64055230
|
2.105
|
NM_177628
|
Fam167a
|
family with sequence similarity 167, member A
|
|
chr4_+_124419115
|
2.103
|
NM_008385
|
Inpp5b
|
inositol polyphosphate-5-phosphatase B
|
|
chr5_-_148118681
|
2.102
|
NM_007673
|
Cdx2
|
caudal type homeobox 2
|
|
chr4_+_59202363
|
2.081
|
NM_011673
|
Ugcg
|
UDP-glucose ceramide glucosyltransferase
|
|
chr6_+_71657791
|
2.071
|
NM_178608
|
Reep1
|
receptor accessory protein 1
|
|
chr1_-_64784316
|
2.063
|
NM_001042659
NM_022721
|
Fzd5
|
frizzled homolog 5 (Drosophila)
|