|
chr11_-_90252228
|
17.553
|
NM_172563
|
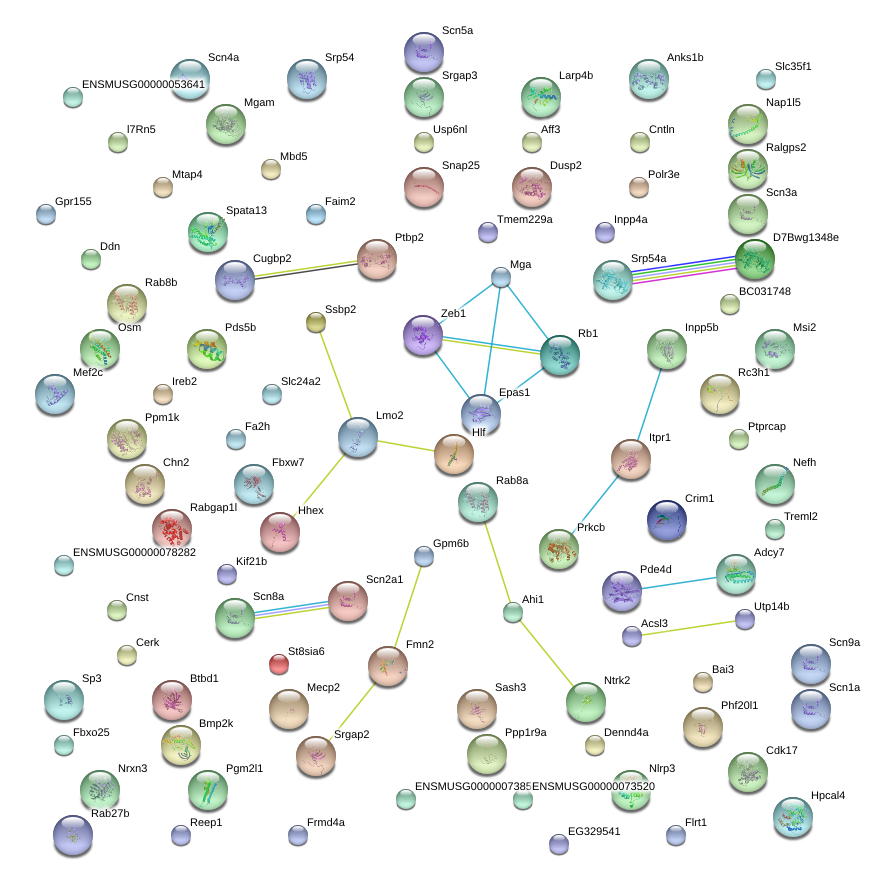
Hlf
|
hepatic leukemia factor
|
|
chr2_+_103809709
|
12.589
|
NM_001142336
|
Lmo2
|
LIM domain only 2
|
|
chr2_+_103798150
|
11.472
|
NM_001142337
|
Lmo2
|
LIM domain only 2
|
|
chr7_+_129432585
|
11.303
|
NM_008855
|
Prkcb
|
protein kinase C, beta
|
|
chr7_+_107376116
|
11.017
|
NM_027629
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr2_+_103810274
|
10.305
|
NM_001142335
NM_008505
|
Lmo2
|
LIM domain only 2
|
|
chr19_+_37509290
|
9.656
|
NM_008245
|
Hhex
|
hematopoietically expressed homeobox
|
|
chr1_+_37356722
|
9.581
|
NM_030266
|
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I
|
|
chr10_+_92623587
|
8.694
|
NM_146239
|
Cdk17
|
cyclin-dependent kinase 17
|
|
chr1_+_162836541
|
8.265
|
NM_001024952
|
Rc3h1
|
RING CCCH (C3H) domains 1
|
|
chr3_+_84619399
|
8.135
|
NM_001177773
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr3_+_84668066
|
7.707
|
NM_001177774
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr2_+_127161893
|
6.833
|
NM_010090
|
Dusp2
|
dual specificity phosphatase 2
|
|
chr9_-_66767436
|
6.699
|
NM_173413
|
Rab8b
|
RAB8B, member RAS oncogene family
|
|
chr3_-_119486305
|
6.646
|
NM_019550
|
Ptbp2
|
polypyrimidine tract binding protein 2
|
|
chr2_-_7002344
|
6.511
|
NM_001110229
NM_001110230
NM_001160292
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr2_-_73224394
|
6.274
|
NM_001080707
NM_001190297
|
Gpr155
|
G protein-coupled receptor 155
|
|
chr4_-_86876328
|
6.142
|
NM_172426
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr10_+_89336253
|
6.067
|
NM_001128086
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr4_+_122860705
|
5.815
|
NM_174998
|
Hpcal4
|
hippocalcin-like 4
|
|
chr5_+_97426665
|
5.696
|
NM_080708
|
Bmp2k
|
BMP2 inducible kinase
|
|
chr2_-_66278870
|
5.522
|
NM_018733
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha
|
|
chr2_-_7317824
|
5.264
|
NM_001110231
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr2_-_7317513
|
5.220
|
NM_001110232
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_65508501
|
5.181
|
NM_001099298
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1
|
|
chr9_+_109834266
|
5.123
|
NM_008633
|
Mtap4
|
microtubule-associated protein 4
|
|
chr8_+_90818102
|
5.059
|
NM_001037724
|
Adcy7
|
adenylate cyclase 7
|
|
chr2_+_4480799
|
5.036
|
NM_001177844
|
Frmd4a
|
FERM domain containing 4A
|
|
chr8_+_90812964
|
4.944
|
NM_001037723
|
Adcy7
|
adenylate cyclase 7
|
|
chr2_+_136539143
|
4.872
|
NM_011428
|
Snap25
|
synaptosomal-associated protein 25
|
|
chr1_+_176431933
|
4.822
|
NM_019445
|
Fmn2
|
formin 2
|
|
chr4_+_124419115
|
4.801
|
NM_008385
|
Inpp5b
|
inositol polyphosphate-5-phosphatase B
|
|
chr15_-_98638348
|
4.770
|
NM_001013741
|
Ddn
|
dendrin
|
|
chr8_+_90806890
|
4.760
|
NM_001109756
|
Adcy7
|
adenylate cyclase 7
|
|
chr6_+_65621583
|
4.729
|
NM_172399
|
A930038C07Rik
|
RIKEN cDNA A930038C07 gene
|
|
chr5_+_151476361
|
4.707
|
NM_175310
|
Pds5b
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr1_-_25886551
|
4.634
|
NM_175642
|
Bai3
|
brain-specific angiogenesis inhibitor 3
|
|
chrX_+_45499599
|
4.618
|
NM_028773
|
Sash3
|
SAM and SH3 domain containing 3
|
|
chr8_+_13907735
|
4.561
|
NM_025785
|
Fbxo25
|
F-box protein 25
|
|
chr2_-_6805937
|
4.433
|
NM_001110228
NM_001160293
NM_010160
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr14_+_61253517
|
4.432
|
NM_001033272
|
Spata13
|
spermatogenesis associated 13
|
|
chr6_-_112897259
|
4.413
|
NM_080448
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr6_-_57485419
|
4.391
|
NM_175523
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing)
|
|
chr2_+_6243767
|
4.390
|
NM_001080548
|
Usp6nl
|
USP6 N-terminal like
|
|
chr10_+_90039435
|
4.375
|
NM_001177396
NM_001177398
NM_181398
|
Ipw
Anks1b
|
imprinted gene in the Prader-Willi syndrome region
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr10_+_90038471
|
4.335
|
NM_001177397
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr17_+_78599556
|
4.316
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr6_+_53989925
|
4.301
|
NM_001163640
|
Chn2
|
chimerin (chimaerin) 2
|
|
chr10_+_52410306
|
4.286
|
NM_178675
|
Slc35f1
|
solute carrier family 35, member F1
|
|
chr8_+_90796301
|
4.283
|
NM_007406
|
Adcy7
|
adenylate cyclase 7
|
|
chr2_+_6273778
|
4.258
|
NM_181399
|
Usp6nl
|
USP6 N-terminal like
|
|
chr6_-_58857000
|
4.246
|
NM_021432
|
Nap1l5
|
nucleosome assembly protein 1-like 5
|
|
chr13_+_83643032
|
4.225
|
NM_001170537
NM_025282
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr2_-_65405546
|
4.222
|
NM_018732
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha
|
|
chr11_-_4847968
|
4.167
|
NM_010904
|
Nefh
|
neurofilament, heavy polypeptide
|
|
chr11_-_88579517
|
4.164
|
NM_054043
|
Msi2
|
Musashi homolog 2 (Drosophila)
|
|
chr2_+_4073908
|
4.097
|
NM_172475
|
Frmd4a
|
FERM domain containing 4A
|
|
chr18_+_5591869
|
4.083
|
NM_011546
|
Zeb1
|
zinc finger E-box binding homeobox 1
|
|
chr2_-_104250490
|
4.078
|
NM_001033347
|
D430041D05Rik
|
RIKEN cDNA D430041D05 gene
|
|
chr1_-_158869401
|
4.046
|
NM_001159966
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr9_+_64658817
|
4.010
|
NM_001162917
|
Dennd4a
|
DENN/MADD domain containing 4A
|
|
chr6_-_24905939
|
4.000
|
NM_177013
|
Tmem229a
|
transmembrane protein 229A
|
|
chr13_+_109444370
|
3.945
|
NM_011056
|
Pde4d
|
phosphodiesterase 4D, cAMP specific
|
|
chr15_+_66409094
|
3.942
|
NM_001081409
|
Phf20l1
|
PHD finger protein 20-like 1
|
|
chr12_+_56256090
|
3.892
|
NM_001100109
|
Srp54b
|
signal recognition particle 54B
|
|
chr11_+_59356187
|
3.847
|
NM_145827
|
Nlrp3
|
NLR family, pyrin domain containing 3
|
|
chr2_+_4322020
|
3.792
|
NM_001177843
|
Frmd4a
|
FERM domain containing 4A
|
|
chr2_-_72818502
|
3.755
|
NM_001018042
|
Sp3
|
trans-acting transcription factor 3
|
|
chr2_+_48805027
|
3.683
|
NM_029924
|
Mbd5
|
methyl-CpG binding domain protein 5
|
|
chr18_-_70212948
|
3.652
|
NM_030554
|
Rab27b
|
RAB27b, member RAS oncogene family
|
|
chr6_+_54222814
|
3.602
|
NM_023543
|
Chn2
|
chimerin (chimaerin) 2
|
|
chr4_-_86873866
|
3.579
|
NM_001110240
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr13_+_91600662
|
3.448
|
NM_024186
NM_024272
|
Ssbp2
|
single-stranded DNA binding protein 2
|
|
chr2_+_119722957
|
3.416
|
NM_001164274
NM_013720
|
Mga
|
MAX gene associated
|
|
chr2_-_13715101
|
3.364
|
NM_145838
|
St8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6
|
|
chr12_+_90191833
|
3.304
|
NM_172544
|
Nrxn3
|
neurexin III
|
|
chr19_+_4154645
|
3.291
|
NM_016933
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein
|
|
chr18_-_70301202
|
3.275
|
NM_001082553
|
Rab27b
|
RAB27b, member RAS oncogene family
|
|
chr6_+_108163089
|
3.274
|
NM_010585
|
Itpr1
|
inositol 1,4,5-triphosphate receptor 1
|
|
chr7_-_88974235
|
3.232
|
NM_146193
|
Btbd1
|
BTB (POZ) domain containing 1
|
|
chr6_+_71657791
|
3.152
|
NM_178608
|
Reep1
|
receptor accessory protein 1
|
|
chr19_-_7180162
|
3.145
|
NM_201411
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr14_-_73725576
|
3.126
|
NM_009029
|
Rb1
|
retinoblastoma 1
|
|
chr15_-_86016336
|
3.092
|
NM_145475
|
Cerk
|
ceramide kinase
|
|
chrX_+_162676870
|
3.068
|
NM_001177961
NM_001177962
NM_023122
|
Gpm6b
|
glycoprotein m6b
|
|
chr17_+_48439264
|
3.053
|
NM_001033405
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2
|
|
chr13_+_9093121
|
3.023
|
NM_172585
|
Larp4b
|
La ribonucleoprotein domain family, member 4B
|
|
chr7_+_128061257
|
3.023
|
NM_001164096
NM_025298
|
Polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E
|
|
chr13_+_58907953
|
3.019
|
NM_008745
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr1_-_158869650
|
3.017
|
NM_001159965
NM_001159967
NM_001159968
NM_023884
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr10_+_20760625
|
2.993
|
NM_001177776
|
Ahi1
|
Abelson helper integration site 1
|
|
chrX_+_162782484
|
2.978
|
NM_001177955
NM_001177956
NM_001177957
NM_001177958
NM_001177959
NM_001177960
|
Gpm6b
|
glycoprotein m6b
|
|
chrX_+_133584130
|
2.968
|
NM_146261
|
Fam199x
|
family with sequence similarity 199, X-linked
|
|
chr1_-_162281701
|
2.957
|
NM_001038621
|
Rabgap1l
|
RAB GTPase activating protein 1-like
|
|
chr6_+_4853319
|
2.944
|
NM_181595
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr8_-_113917651
|
2.942
|
NM_178086
|
Fa2h
|
fatty acid 2-hydroxylase
|
|
chr1_+_78654399
|
2.903
|
NM_001001981
NM_001033606
NM_001136222
NM_028817
|
Utp14b
Acsl3
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog B (yeast)
acyl-CoA synthetase long-chain family member 3
|
|
chr4_+_84530230
|
2.902
|
NM_175275
NM_177385
|
Cntln
|
centlein, centrosomal protein
|
|
chr1_+_138027943
|
2.892
|
NM_001039472
|
Kif21b
|
kinesin family member 21B
|
|
chr1_+_181476534
|
2.873
|
NM_146105
|
Cnst
|
consortin, connexin sorting protein
|
|
chr1_-_38721799
|
2.802
|
NM_010678
|
Aff3
|
AF4/FMR2 family, member 3
|
|
chr15_-_99358387
|
2.743
|
NM_001038658
NM_028224
|
Faim2
|
Fas apoptotic inhibitory molecule 2
|
|
chr11_+_4136787
|
2.741
|
NM_001013365
|
Osm
|
oncostatin M
|
|
chr13_-_94269616
|
2.695
|
NM_021310
|
Jmy
|
junction-mediating and regulatory protein
|
|
chr12_-_71448600
|
2.691
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr14_+_120877548
|
2.686
|
NM_029519
|
Rap2a
|
RAS related protein 2a
|
|
chr5_+_22252006
|
2.667
|
NM_001081231
NM_029990
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3
|
|
chr16_-_78576898
|
2.630
|
NM_025967
|
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed
|
|
chr2_-_51790163
|
2.627
|
NM_001141982
|
Rbm43
|
RNA binding motif protein 43
|
|
chr16_+_17489743
|
2.607
|
NM_175178
|
Aifm3
|
apoptosis-inducing factor, mitochondrion-associated 3
|
|
chr2_-_51790406
|
2.544
|
NM_001141981
NM_030243
|
Rbm43
|
RNA binding motif protein 43
|
|
chr5_+_35104373
|
2.544
|
NM_010414
|
Htt
|
huntingtin
|
|
chr15_-_84388245
|
2.540
|
NM_177630
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr1_-_74590801
|
2.536
|
NM_176972
|
Usp37
|
ubiquitin specific peptidase 37
|
|
chr3_-_36589048
|
2.534
|
NM_019510
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr2_+_49921978
|
2.525
|
NM_177139
|
Lypd6
|
LY6/PLAUR domain containing 6
|
|
chr8_+_90723069
|
2.506
|
NM_001164497
NM_001164498
NM_001164499
|
Papd5
|
PAP associated domain containing 5
|
|
chr9_-_54509658
|
2.496
|
NM_053178
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr4_+_138011061
|
2.478
|
NM_025451
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr16_+_13358513
|
2.454
|
NM_153588
|
Mkl2
|
MKL/myocardin-like 2
|
|
chr6_-_28781687
|
2.431
|
NM_138682
|
Lrrc4
|
leucine rich repeat containing 4
|
|
chr15_+_99532698
|
2.408
|
NM_031842
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr18_-_66129715
|
2.403
|
NM_145493
|
Cplx4
|
complexin 4
|
|
chr13_-_111070392
|
2.371
|
NM_023852
|
Rab3c
|
RAB3C, member RAS oncogene family
|
|
chr1_+_132613903
|
2.349
|
NM_178691
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae)
|
|
chr1_-_171677653
|
2.306
|
NM_009062
|
Rgs4
|
regulator of G-protein signaling 4
|
|
chr8_-_63462080
|
2.296
|
NM_007711
NM_173873
NM_173874
|
Clcn3
|
chloride channel 3
|
|
chr5_-_100588826
|
2.289
|
NM_182841
|
Tmem150c
|
transmembrane protein 150C
|
|
chr2_-_136213509
|
2.287
|
NM_172858
|
Pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr2_+_28048592
|
2.285
|
NM_001038613
NM_001038614
|
Olfm1
|
olfactomedin 1
|
|
chr19_+_12873063
|
2.282
|
NM_134152
|
Lpxn
|
leupaxin
|
|
chr12_-_4038848
|
2.267
|
NM_001082483
|
Efr3b
|
EFR3 homolog B (S. cerevisiae)
|
|
chr7_+_107855215
|
2.256
|
NM_178764
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr5_-_87174825
|
2.248
|
NM_172880
|
Tmprss11e
|
transmembrane protease, serine 11e
|
|
chr1_+_148342795
|
2.240
|
NM_153539
|
Fam5c
|
family with sequence similarity 5, member C
|
|
chr6_+_134590957
|
2.204
|
NM_026371
|
Loh12cr1
|
loss of heterozygosity, 12, chromosomal region 1 homolog (human)
|
|
chr2_+_13995715
|
2.202
|
NM_011484
|
Stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1
|
|
chr10_-_88720534
|
2.174
|
NM_178773
|
Ano4
|
anoctamin 4
|
|
chr5_+_123300153
|
2.173
|
NM_030564
|
Rnf34
|
ring finger protein 34
|
|
chr6_+_86354212
|
2.168
|
NM_001164078
NM_001164079
NM_011585
|
Tia1
|
cytotoxic granule-associated RNA binding protein 1
|
|
chr15_+_81767188
|
2.145
|
NM_145473
|
Csdc2
|
cold shock domain containing C2, RNA binding
|
|
chr19_-_19185685
|
2.145
|
NM_001043354
|
Rorb
|
RAR-related orphan receptor beta
|
|
chr10_+_20672352
|
2.142
|
NM_026203
|
Ahi1
|
Abelson helper integration site 1
|
|
chr2_-_72817311
|
2.141
|
NM_001098425
|
Sp3
|
trans-acting transcription factor 3
|
|
chr6_-_91465788
|
2.140
|
NM_009531
|
Xpc
|
xeroderma pigmentosum, complementation group C
|
|
chr13_+_41701584
|
2.127
|
NM_001163572
|
Tmem170b
|
transmembrane protein 170B
|
|
chr6_+_125095236
|
2.126
|
NM_178787
NM_001039669
|
Iffo1
|
intermediate filament family orphan 1
|
|
chrX_+_83115611
|
2.118
|
NM_001163539
|
5430427O19Rik
|
RIKEN cDNA 5430427O19 gene
|
|
chr5_-_25004598
|
2.115
|
NM_001081383
|
Mll3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr3_-_9833502
|
2.099
|
NM_001195031
NM_053182
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr8_-_63433528
|
2.097
|
NM_173876
|
Clcn3
|
chloride channel 3
|
|
chrX_+_103222548
|
2.096
|
NM_001109757
NM_009726
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr5_+_30559138
|
2.089
|
NM_027652
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr1_+_108068405
|
2.074
|
NM_133821
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr4_+_154338216
|
2.066
|
NM_172990
|
Pank4
|
pantothenate kinase 4
|
|
chr4_+_154624074
|
2.058
|
NM_001190445
|
2610002J02Rik
|
RIKEN cDNA 2610002J02 gene
|
|
chr8_+_23970006
|
2.038
|
NM_001081149
|
Myst3
|
MYST histone acetyltransferase (monocytic leukemia) 3
|
|
chr13_-_14615371
|
2.035
|
NM_001081348
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr18_+_42435006
|
2.031
|
NM_172626
|
Rbm27
|
RNA binding motif protein 27
|
|
chr13_-_52626065
|
2.017
|
NM_001024474
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr5_-_100468240
|
2.015
|
NM_016690
|
Hnrpdl
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr13_+_93195850
|
2.006
|
NM_175455
|
Ankrd34b
|
ankyrin repeat domain 34B
|
|
chr18_+_33098694
|
1.993
|
NM_009793
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr5_-_118694869
|
1.989
|
NM_172998
NM_001109902
|
Rnft2
|
ring finger protein, transmembrane 2
|
|
chr1_+_129670740
|
1.988
|
NM_028399
|
Ccnt2
|
cyclin T2
|
|
chr11_+_93857338
|
1.959
|
NM_027569
|
Spag9
|
sperm associated antigen 9
|
|
chr2_+_31428244
|
1.952
|
NM_001033389
|
Fubp3
|
far upstream element (FUSE) binding protein 3
|
|
chr2_+_158235580
|
1.944
|
NM_177658
|
Ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr11_+_68902025
|
1.943
|
NM_009497
|
Vamp2
|
vesicle-associated membrane protein 2
|
|
chr15_+_6658368
|
1.940
|
NM_030168
|
Rictor
|
RPTOR independent companion of MTOR, complex 2
|
|
chr1_-_94998418
|
1.920
|
NM_001110315
NM_008440
|
Kif1a
|
kinesin family member 1A
|
|
chr7_-_88837850
|
1.918
|
NM_001164087
|
Homer2
|
homer homolog 2 (Drosophila)
|
|
chr17_+_47748530
|
1.910
|
NM_020048
|
Med20
|
mediator complex subunit 20
|
|
chr2_+_28061156
|
1.901
|
NM_001038612
NM_019498
|
Olfm1
|
olfactomedin 1
|
|
chr9_-_123883398
|
1.895
|
NM_009912
|
Ccr1
|
chemokine (C-C motif) receptor 1
|
|
chrX_+_97972559
|
1.892
|
NM_001177780
|
Dlg3
|
discs, large homolog 3 (Drosophila)
|
|
chr1_+_148344816
|
1.888
|
NM_001145807
|
Fam5c
|
family with sequence similarity 5, member C
|
|
chr7_+_63023547
|
1.887
|
NM_178706
|
Siglech
|
sialic acid binding Ig-like lectin H
|
|
chr9_+_104472090
|
1.878
|
NM_028719
|
Cpne4
|
copine IV
|
|
chr3_-_30868320
|
1.868
|
NM_001165954
NM_001165955
NM_001165956
NM_153421
|
Phc3
|
polyhomeotic-like 3 (Drosophila)
|
|
chr7_+_91040481
|
1.842
|
NM_023403
|
Mesdc2
|
mesoderm development candidate 2
|
|
chrX_+_97963061
|
1.841
|
NM_001177778
NM_016747
|
Dlg3
|
discs, large homolog 3 (Drosophila)
|
|
chr17_+_26852581
|
1.840
|
NM_029870
|
A930001N09Rik
|
RIKEN cDNA A930001N09 gene
|
|
chr10_+_122701051
|
1.837
|
NM_182807
|
Fam19a2
|
family with sequence similarity 19, member A2
|
|
chr15_+_81633102
|
1.833
|
NM_153484
|
Tef
|
thyrotroph embryonic factor
|
|
chr5_-_90312358
|
1.831
|
NM_001081401
NM_177872
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3
|
|
chr11_-_61668486
|
1.786
|
NM_013881
|
Ulk2
|
Unc-51 like kinase 2 (C. elegans)
|
|
chr12_+_89961317
|
1.782
|
NM_001198587
|
Nrxn3
|
neurexin III
|
|
chr6_-_77929660
|
1.778
|
NM_001109764
NM_009819
NM_145732
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr7_+_104480819
|
1.766
|
NM_001039039
|
Kctd21
|
potassium channel tetramerisation domain containing 21
|
|
chr12_+_112088488
|
1.760
|
NM_028365
|
Zfp839
|
zinc finger protein 839
|
|
chr9_+_66561492
|
1.760
|
NM_178396
|
Car12
|
carbonic anyhydrase 12
|
|
chr1_-_173164404
|
1.720
|
NM_010185
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide
|
|
chr16_+_45610535
|
1.718
|
NM_001159297
NM_008099
|
Gcet2
|
germinal center expressed transcript 2
|
|
chr2_-_180444687
|
1.718
|
NM_177852
|
Dido1
|
death inducer-obliterator 1
|
|
chr3_-_87734096
|
1.677
|
NM_153562
|
Rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chr2_-_162486882
|
1.660
|
NM_021464
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T
|
|
chr7_+_134439804
|
1.653
|
NM_008400
|
Itgal
|
integrin alpha L
|
|
chr1_+_179374783
|
1.651
|
NM_001012330
|
Zfp238
|
zinc finger protein 238
|
|
chr6_+_134589529
|
1.635
|
NM_001170479
|
Loh12cr1
|
loss of heterozygosity, 12, chromosomal region 1 homolog (human)
|