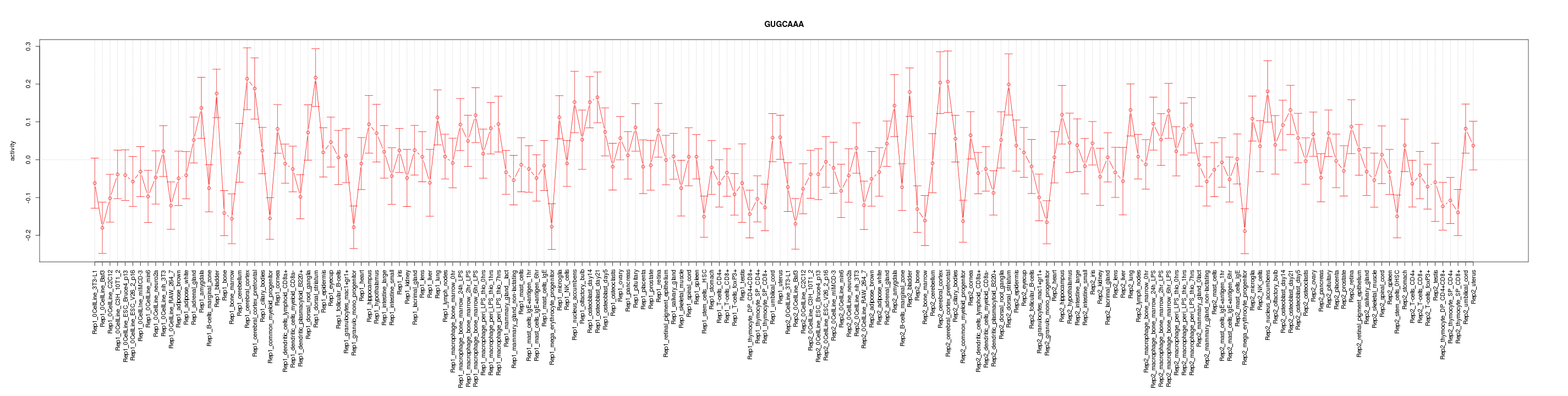
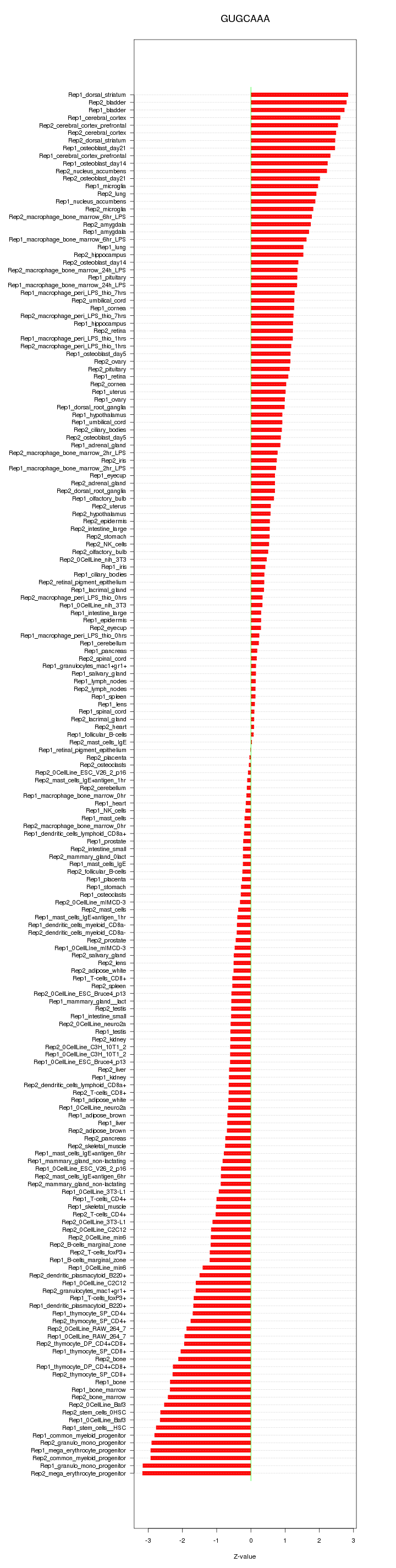
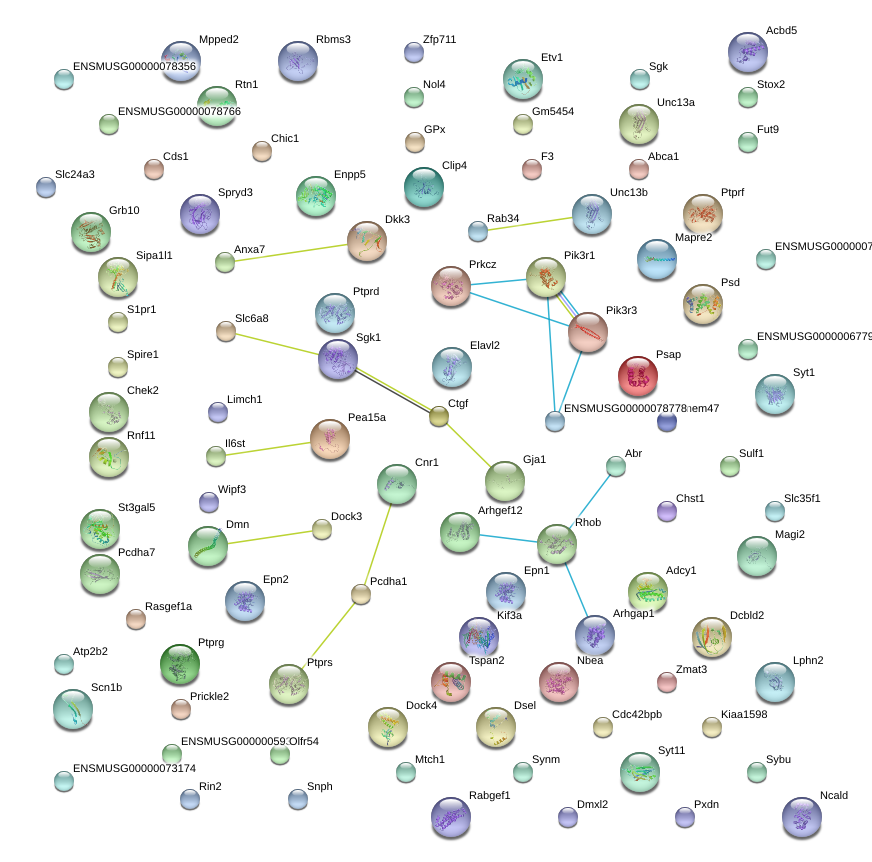
|
chr6_-_92517359
|
23.719
|
NM_001134461
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr6_-_92656148
|
22.675
|
NM_001081146
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr6_-_92484838
|
22.629
|
NM_001134459
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr6_-_92431385
|
21.504
|
NM_001134460
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr12_-_73337707
|
19.938
|
NM_001007596
|
Rtn1
|
reticulon 1
|
|
chr12_-_73509925
|
19.747
|
NM_153457
|
Rtn1
|
reticulon 1
|
|
chr7_-_119302546
|
16.096
|
NM_015814
|
Dkk3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr3_-_55987612
|
16.022
|
NM_030595
|
Nbea
|
neurobeachin
|
|
chrX_+_78315963
|
14.958
|
NM_138751
|
Tmem47
|
transmembrane protein 47
|
|
chr3_-_88576415
|
14.489
|
NM_018804
|
Syt11
|
synaptotagmin XI
|
|
chr3_-_148617598
|
14.443
|
NM_001081298
|
Lphn2
|
latrophilin 2
|
|
chr4_+_115894518
|
13.765
|
NM_181585
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55)
|
|
chr8_-_48437701
|
13.637
|
NM_175162
|
Stox2
|
storkhead box 2
|
|
chr10_+_24315247
|
13.495
|
NM_010217
|
Ctgf
|
connective tissue growth factor
|
|
chr6_+_54402876
|
13.366
|
NM_001167860
NM_001167861
|
Wipf3
|
WAS/WASL interacting protein family, member 3
|
|
chr8_-_48374755
|
13.210
|
NM_001114311
|
Stox2
|
storkhead box 2
|
|
chr12_-_8506790
|
12.952
|
NM_007483
|
Rhob
|
ras homolog gene family, member B
|
|
chr19_-_59150558
|
12.700
|
NM_001114312
NM_175172
|
4930506M07Rik
|
RIKEN cDNA 4930506M07 gene
|
|
chr17_+_44215761
|
12.598
|
NM_032003
NM_001168620
|
Enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5
|
|
chr9_-_117539030
|
12.402
|
NM_001172121
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr12_+_30622876
|
12.327
|
NM_181395
|
Pxdn
|
peroxidasin homolog (Drosophila)
|
|
chr1_+_12682510
|
12.300
|
NM_001198565
NM_001198566
|
Sulf1
|
sulfatase 1
|
|
chr1_-_113761344
|
12.214
|
NM_001081316
|
Dsel
|
dermatan sulfate epimerase-like
|
|
chr6_+_118016385
|
12.100
|
NM_027526
|
Rasgef1a
|
RasGEF domain family, member 1A
|
|
chr9_-_117161564
|
12.017
|
NM_001172122
NM_001172123
NM_001172124
NM_001172126
NM_178660
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr2_+_92439758
|
11.937
|
NM_023850
|
Chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr9_-_107134182
|
11.901
|
NM_153413
|
Dock3
|
dedicator of cyto-kinesis 3
|
|
chr1_-_174136874
|
11.604
|
NM_011063
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A
|
|
chr6_-_113841356
|
11.454
|
NM_001036684
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr15_-_37722161
|
11.307
|
NM_134094
|
Ncald
|
neurocalcin delta
|
|
chr10_+_59747208
|
11.268
|
NM_001146124
|
Psap
|
prosaposin
|
|
chr5_+_67137054
|
11.183
|
NM_001001980
|
Limch1
|
LIM and calponin homology domains 1
|
|
chr2_+_145068346
|
10.907
|
NM_053195
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr4_-_25727109
|
10.763
|
NM_010243
|
Fut9
|
fucosyltransferase 9
|
|
chr15_-_37721754
|
10.696
|
NM_001170866
|
Ncald
|
neurocalcin delta
|
|
chr2_+_91490266
|
10.664
|
NM_001145902
|
Arhgap1
|
Rho GTPase activating protein 1
|
|
chr19_-_46401618
|
10.645
|
NM_028627
|
Psd
|
pleckstrin and Sec7 domain containing
|
|
chr6_-_113991945
|
10.477
|
NM_009723
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr1_+_12708582
|
10.473
|
NM_172294
|
Sulf1
|
sulfatase 1
|
|
chr10_+_59740373
|
10.312
|
NM_001146120
NM_001146121
NM_001146122
NM_001146123
NM_011179
|
Psap
|
prosaposin
|
|
chr15_-_44619541
|
10.127
|
NM_001032727
NM_176998
|
Sybu
|
syntabulin (syntaxin-interacting)
|
|
chr2_+_145611851
|
10.063
|
NM_028724
|
Rin2
|
Ras and Rab interactor 2
|
|
chr12_+_83412335
|
10.016
|
NM_172579
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1
|
|
chr12_+_41172639
|
9.776
|
NM_172803
|
Dock4
|
dedicator of cytokinesis 4
|
|
chrX_+_70918446
|
9.741
|
NM_001142809
NM_001142810
NM_133987
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr4_-_76240202
|
9.566
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr15_-_44584003
|
9.514
|
NM_178765
|
Sybu
|
syntabulin (syntaxin-interacting)
|
|
chr11_+_53380870
|
9.499
|
NM_008443
|
Kif3a
|
kinesin family member 3A
|
|
chr18_+_4634926
|
9.123
|
NM_001081963
|
9430020K01Rik
|
RIKEN cDNA 9430020K01 gene
|
|
chr3_-_115417932
|
9.068
|
NM_007901
|
S1pr1
|
sphingosine-1-phosphate receptor 1
|
|
chr10_+_56097078
|
9.021
|
NM_010288
|
Gja1
|
gap junction protein, alpha 1
|
|
chr15_-_37389073
|
8.927
|
NM_001170867
NM_001170868
|
Ncald
|
neurocalcin delta
|
|
chr18_-_23197156
|
8.833
|
NM_001161483
|
Nol4
|
nucleolar protein 4
|
|
chr3_+_121426422
|
8.823
|
NM_010171
|
F3
|
coagulation factor III
|
|
chr4_-_154735351
|
8.770
|
NM_008860
|
Prkcz
|
protein kinase C, zeta
|
|
chr10_+_21712008
|
8.725
|
NM_001161849
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr9_-_54349409
|
8.696
|
NM_172771
|
Dmxl2
|
Dmx-like 2
|
|
chr18_+_37089837
|
8.582
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chr2_-_151458256
|
8.559
|
NM_198214
|
Snph
|
syntaphilin
|
|
chr5_+_102194126
|
8.534
|
NM_173370
|
Cds1
|
CDP-diacylglycerol synthase 1
|
|
chr18_+_37120093
|
8.531
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr18_+_37179802
|
8.463
|
NM_138663
|
Pcdha12
|
protocadherin alpha 12
|
|
chr18_+_37098858
|
8.442
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr11_-_11927973
|
8.425
|
NM_001177629
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr4_-_75779379
|
8.408
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_+_21698470
|
8.345
|
NM_001161847
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_+_12386043
|
8.316
|
NM_008981
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G
|
|
chr10_-_108447674
|
8.313
|
NM_009306
|
Syt1
|
synaptotagmin I
|
|
chr18_+_37157533
|
8.298
|
NM_138661
|
Pcdha9
|
protocadherin alpha 9
|
|
chr2_+_106535757
|
8.155
|
NM_029837
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr3_-_32264572
|
8.127
|
NM_009517
|
Zmat3
|
zinc finger matrin type 3
|
|
chr18_+_37127284
|
7.990
|
NM_007767
|
Pcdha6
|
protocadherin alpha 6
|
|
chr11_-_61393108
|
7.908
|
NM_010148
|
Epn2
|
epsin 2
|
|
chr3_+_102538692
|
7.881
|
NM_027533
|
Tspan2
|
tetraspanin 2
|
|
chr18_+_37105758
|
7.860
|
NM_138662
|
Pcdha3
|
protocadherin alpha 3
|
|
chr10_+_21705909
|
7.774
|
NM_001161848
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chrX_+_100551808
|
7.745
|
NM_009767
|
Chic1
|
cysteine-rich hydrophobic domain 1
|
|
chr8_-_74195650
|
7.666
|
NM_001029873
|
Unc13a
|
unc-13 homolog A (C. elegans)
|
|
chr4_-_53172748
|
7.604
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr10_+_21714471
|
7.551
|
NM_011361
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr18_+_37152024
|
7.463
|
NM_201243
|
Pcdha8
|
protocadherin alpha 8
|
|
chr18_+_37133455
|
7.453
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chr7_-_31911884
|
7.386
|
NM_011322
|
Scn1b
|
sodium channel, voltage-gated, type I, beta
|
|
chr6_+_72047563
|
7.320
|
NM_001035228
NM_011375
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr4_-_91038729
|
7.309
|
NM_010486
NM_207686
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr10_+_21601968
|
7.036
|
NM_001161845
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_+_21713669
|
6.867
|
NM_001161850
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr4_+_34011567
|
6.816
|
NM_007726
|
Cnr1
|
cannabinoid receptor 1 (brain)
|
|
chr10_+_52410306
|
6.801
|
NM_178675
|
Slc35f1
|
solute carrier family 35, member F1
|
|
chr2_+_106533615
|
6.789
|
NM_001143683
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr4_-_154719834
|
6.764
|
NM_001039079
|
Prkcz
|
protein kinase C, zeta
|
|
chr9_-_42913797
|
6.759
|
NM_027144
|
Arhgef12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chrX_+_109714134
|
6.732
|
NM_177747
|
Zfp711
|
zinc finger protein 711
|
|
chr12_+_83271002
|
6.623
|
NM_001167983
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1
|
|
chr4_-_91042997
|
6.617
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr11_+_6963421
|
6.542
|
NM_009622
|
Adcy1
|
adenylate cyclase 1
|
|
chr12_-_112615921
|
6.451
|
NM_183016
|
Cdc42bpb
|
CDC42 binding protein kinase beta
|
|
chr17_+_72131286
|
6.396
|
NM_030179
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr11_+_78002249
|
6.382
|
NM_033475
|
Rab34
|
RAB34, member of RAS oncogene family
|
|
chr2_+_22923653
|
6.373
|
NM_001102436
NM_028793
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5
|
|
chr13_+_113254277
|
6.343
|
NM_010560
|
Il6st
|
interleukin 6 signal transducer
|
|
chr4_-_109149038
|
6.341
|
NM_013876
|
Rnf11
|
ring finger protein 11
|
|
chr11_-_76322920
|
6.322
|
NM_198018
|
Abr
|
active BCR-related gene
|
|
chr11_+_78001871
|
6.271
|
NM_001159482
|
Rab34
|
RAB34, member of RAS oncogene family
|
|
chr5_+_18732863
|
6.257
|
NM_001170746
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr15_-_101966611
|
6.242
|
NM_001033277
|
Spryd3
|
SPRY domain containing 3
|
|
chr12_+_39506844
|
6.191
|
NM_007960
|
Etv1
|
ets variant gene 1
|
|
chr16_+_58408609
|
6.157
|
NM_028523
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr7_-_74904546
|
6.121
|
NM_183312
NM_201639
NM_207663
|
Synm
|
synemin, intermediate filament protein
|
|
chr18_-_67708826
|
6.069
|
NM_176832
|
Spire1
|
spire homolog 1 (Drosophila)
|
|
chr14_-_21299301
|
6.017
|
NM_001110794
NM_009674
|
Anxa7
|
annexin A7
|
|
chr18_+_37303622
|
5.983
|
NM_001003672
|
Pcdhac2
|
protocadherin alpha subfamily C, 2
|
|
chr11_-_76391228
|
5.905
|
NM_198894
|
Abr
|
active BCR-related gene
|
|
chr18_+_23912225
|
5.809
|
NM_001162941
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr17_-_66799086
|
5.798
|
NM_001114098
NM_172963
|
1110012J17Rik
|
RIKEN cDNA 1110012J17 gene
|
|
chr4_-_91066674
|
5.792
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr14_+_120674886
|
5.787
|
NM_175341
NM_207515
|
Mbnl2
|
muscleblind-like 2
|
|
chr13_-_13485585
|
5.765
|
NM_031999
|
Gpr137b
|
G protein-coupled receptor 137B
|
|
chr8_+_87560809
|
5.750
|
NM_145390
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b)
|
|
chr7_+_127245860
|
5.711
|
NM_145586
|
Tmem159
|
transmembrane protein 159
|
|
chr13_+_94077988
|
5.669
|
NM_152134
|
Homer1
|
homer homolog 1 (Drosophila)
|
|
chr18_+_6765174
|
5.599
|
NM_181070
|
Rab18
|
RAB18, member RAS oncogene family
|
|
chr8_-_70342105
|
5.598
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr4_-_82151180
|
5.597
|
NM_001113209
NM_001113210
NM_008687
|
Nfib
|
nuclear factor I/B
|
|
chr18_+_23910833
|
5.509
|
NM_001162942
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr6_-_72566993
|
5.479
|
NM_009443
|
Tgoln1
|
trans-golgi network protein
|
|
chr13_+_98011057
|
5.472
|
NM_007930
|
Enc1
|
ectodermal-neural cortex 1
|
|
chr6_-_119146426
|
5.458
|
NM_009781
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr5_-_149364353
|
5.444
|
NM_011908
|
Ubl3
|
ubiquitin-like 3
|
|
chr11_-_7113899
|
5.439
|
NM_008343
|
Igfbp3
|
insulin-like growth factor binding protein 3
|
|
chr5_-_76733540
|
5.435
|
NM_007715
|
Clock
|
circadian locomotor output cycles kaput
|
|
chr6_-_119058206
|
5.395
|
NM_001159533
NM_001159534
NM_001159535
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr14_-_66832383
|
5.353
|
NM_001162366
|
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta
|
|
chr18_-_67712374
|
5.352
|
NM_194355
|
Spire1
|
spire homolog 1 (Drosophila)
|
|
chr8_+_124254462
|
5.351
|
NM_020605
|
Jph3
|
junctophilin 3
|
|
chr18_+_23573908
|
5.297
|
NM_010087
NM_207650
|
Dtna
|
dystrobrevin alpha
|
|
chr2_-_173102033
|
5.281
|
NM_022995
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1
|
|
chr8_-_37796104
|
5.260
|
NM_001194941
|
Dlc1
|
deleted in liver cancer 1
|
|
chr6_+_4853319
|
5.241
|
NM_181595
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr13_+_48356795
|
5.110
|
NM_031166
|
Id4
|
inhibitor of DNA binding 4
|
|
chr6_-_12699189
|
5.099
|
NM_001164805
|
Thsd7a
|
thrombospondin, type I, domain containing 7A
|
|
chr4_-_141795288
|
5.039
|
NM_001109685
|
9030409G11Rik
|
RIKEN cDNA 9030409G11 gene
|
|
chr8_-_25549415
|
4.996
|
NM_026931
|
1810011O10Rik
|
RIKEN cDNA 1810011O10 gene
|
|
chr17_+_14416508
|
4.942
|
NM_022315
|
Smoc2
|
SPARC related modular calcium binding 2
|
|
chr12_+_30213224
|
4.941
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr2_+_71819264
|
4.932
|
NM_019688
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr1_+_137662648
|
4.887
|
NM_013750
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr6_-_72566691
|
4.845
|
NM_009444
|
Tgoln1
Tgoln2
|
trans-golgi network protein
trans-golgi network protein 2
|
|
chr8_-_8660610
|
4.844
|
NM_010111
|
Efnb2
|
ephrin B2
|
|
chr16_-_22439642
|
4.831
|
NM_023794
|
Etv5
|
ets variant gene 5
|
|
chr2_-_157392026
|
4.826
|
NM_016916
|
Blcap
|
bladder cancer associated protein homolog (human)
|
|
chr1_-_154472199
|
4.807
|
NM_016846
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1
|
|
chr4_+_13711928
|
4.799
|
NM_009822
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_+_105907394
|
4.797
|
NM_177373
|
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr5_+_19413335
|
4.796
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr10_-_18463500
|
4.664
|
NM_001033258
|
D10Bwg1379e
|
DNA segment, Chr 10, Brigham & Women's Genetics 1379 expressed
|
|
chr19_+_56796861
|
4.657
|
NM_007419
|
Adrb1
|
adrenergic receptor, beta 1
|
|
chr8_-_37676995
|
4.645
|
NM_015802
|
Dlc1
|
deleted in liver cancer 1
|
|
chr15_+_99223377
|
4.582
|
NM_001171034
NM_001171036
NM_026669
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr11_-_86570918
|
4.574
|
NM_001003908
|
Cltc
|
clathrin, heavy polypeptide (Hc)
|
|
chr12_-_25780932
|
4.567
|
NM_010496
|
Id2
|
inhibitor of DNA binding 2
|
|
chr18_+_23962468
|
4.519
|
NM_153058
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr1_+_39251056
|
4.467
|
NM_008719
|
Npas2
|
neuronal PAS domain protein 2
|
|
chr18_+_37112301
|
4.424
|
NM_001174154
NM_007766
|
Pcdha4-g
Pcdha4
|
protocadherin alpha 4-gamma
protocadherin alpha 4
|
|
chr7_+_48154647
|
4.392
|
NM_021387
|
Vstm2b
|
V-set and transmembrane domain containing 2B
|
|
chr15_-_58654964
|
4.329
|
NM_175212
|
Tmem65
|
transmembrane protein 65
|
|
chr17_+_87506971
|
4.326
|
NM_019654
|
Socs5
|
suppressor of cytokine signaling 5
|
|
chr16_+_21204820
|
4.300
|
NM_010143
|
Ephb3
|
Eph receptor B3
|
|
chr2_+_65508501
|
4.300
|
NM_001099298
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1
|
|
chrX_+_53862956
|
4.292
|
NM_172780
|
Slc9a6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr8_+_81552335
|
4.274
|
NM_178267
|
Zfp827
|
zinc finger protein 827
|
|
chr14_-_66899764
|
4.246
|
NM_001162365
NM_172498
|
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta
|
|
chrX_-_20498021
|
4.238
|
NM_001110780
NM_013680
|
Syn1
|
synapsin I
|
|
chr11_-_11937330
|
4.228
|
NM_010345
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr2_+_76244578
|
4.211
|
NM_145525
|
Osbpl6
|
oxysterol binding protein-like 6
|
|
chr15_+_99223622
|
4.196
|
NM_001171035
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr11_+_42233258
|
4.169
|
NM_008070
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2
|
|
chr4_+_13670425
|
4.139
|
NM_001111026
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_30219769
|
4.103
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr7_+_107376116
|
4.092
|
NM_027629
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr7_-_74517624
|
4.085
|
NM_001033713
|
Mef2a
|
myocyte enhancer factor 2A
|
|
chr9_-_16182674
|
4.079
|
NM_001080814
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr16_-_28929698
|
4.044
|
NM_177718
|
1600021P15Rik
|
RIKEN cDNA 1600021P15 gene
|
|
chr11_+_59475986
|
4.013
|
NM_012027
NM_201245
|
Mprip
|
myosin phosphatase Rho interacting protein
|
|
chr12_+_85455277
|
4.011
|
NM_028821
|
Dnalc1
|
dynein, axonemal, light chain 1
|
|
chr2_+_19579607
|
3.964
|
NM_027715
|
Otud1
|
OTU domain containing 1
|
|
chr9_-_105397342
|
3.941
|
NM_175025
|
Atp2c1
|
ATPase, Ca++-sequestering
|
|
chr6_+_86388364
|
3.930
|
NM_146170
|
C87436
|
expressed sequence C87436
|
|
chr2_+_124436031
|
3.929
|
NM_172537
NM_199238
NM_199239
NM_199240
NM_199241
|
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr5_+_111847185
|
3.915
|
NM_001081235
|
Mn1
|
meningioma 1
|
|
chr9_+_58430031
|
3.912
|
NM_009145
|
Nptn
|
neuroplastin
|
|
chr6_+_15135505
|
3.876
|
NM_212435
|
Foxp2
|
forkhead box P2
|
|
chr4_-_151235861
|
3.859
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr2_+_118502976
|
3.783
|
NM_001145854
|
Pak6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr8_-_42140113
|
3.762
|
NM_001005864
|
Mtus1
|
mitochondrial tumor suppressor 1
|
|
chr5_-_53604673
|
3.760
|
NM_172710
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr15_-_39775173
|
3.734
|
NM_172814
|
Lrp12
|
low density lipoprotein-related protein 12
|
|
chr3_+_122432075
|
3.729
|
NM_153422
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific
|
|
chr4_+_97444312
|
3.725
|
NM_001122953
NM_010905
|
Nfia
|
nuclear factor I/A
|
|
chr18_+_3507920
|
3.580
|
NM_026505
|
Bambi
|
BMP and activin membrane-bound inhibitor, homolog (Xenopus laevis)
|