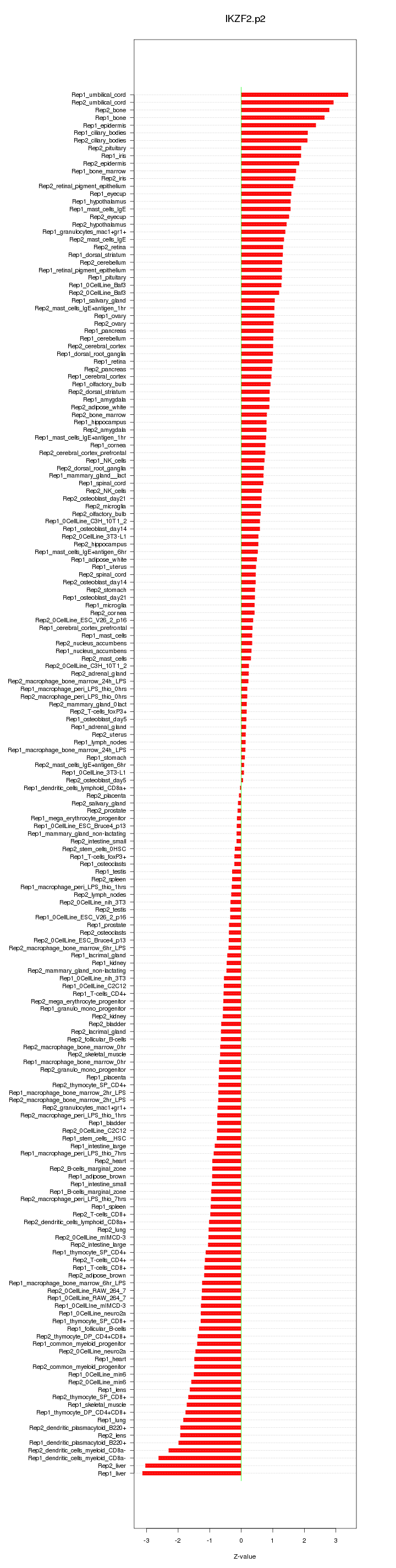
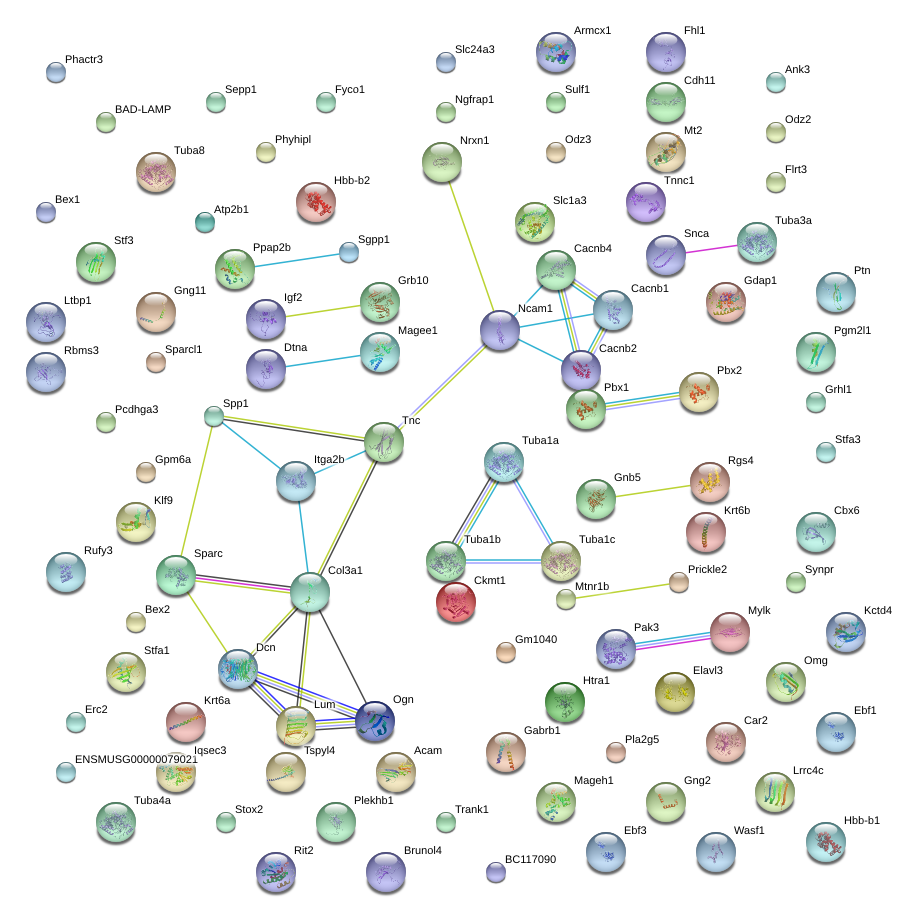
|
chr5_+_72091188
|
20.622
|
NM_008069
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1
|
|
chr8_+_56122680
|
16.029
|
|
Gpm6a
|
glycoprotein m6a
|
|
chrX_+_132806184
|
14.893
|
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr17_-_90436488
|
14.527
|
|
Nrxn1
|
neurexin I
|
|
chr6_-_126658429
|
13.899
|
|
|
|
|
chr9_-_49311823
|
12.943
|
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chrX_+_131252542
|
12.802
|
|
Armcx1
|
armadillo repeat containing, X-linked 1
|
|
chrX_+_131252456
|
12.149
|
NM_001166377
NM_001166378
NM_001166379
NM_001166380
NM_030066
|
Armcx1
|
armadillo repeat containing, X-linked 1
|
|
chr9_-_117160916
|
11.660
|
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr9_-_117161234
|
11.652
|
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr9_-_117161564
|
10.645
|
NM_001172122
NM_001172123
NM_001172124
NM_001172126
NM_178660
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr4_+_104829963
|
9.883
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr4_+_104829867
|
9.273
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr9_-_21820860
|
8.973
|
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr16_-_36334417
|
8.859
|
NM_001001332
|
BC117090
|
cDNA sequence BC1179090
|
|
chr2_+_121183509
|
8.489
|
|
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous
|
|
chr11_-_79317583
|
8.190
|
NM_019409
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr11_-_79317471
|
8.119
|
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr6_+_30756073
|
7.736
|
|
Copg2as2
|
coatomer protein complex, subunit gamma 2, antisense 2
|
|
chr4_+_104829951
|
7.652
|
NM_080555
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_97307652
|
7.051
|
NM_178725
|
Lrrc4c
|
leucine rich repeat containing 4C
|
|
chr2_+_177876635
|
6.722
|
NM_028806
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr11_-_55233354
|
6.651
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr19_+_23241546
|
6.577
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr1_+_45368443
|
6.446
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr6_-_36759929
|
6.370
|
|
Ptn
|
pleiotrophin
|
|
chr2_+_135883963
|
6.304
|
|
6330527O06Rik
|
RIKEN cDNA 6330527O06 gene
|
|
chr7_+_138087002
|
6.265
|
|
Htra1
|
HtrA serine peptidase 1
|
|
chrX_+_140127827
|
6.239
|
NM_001195046
NM_001195047
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chrX_+_10173557
|
6.220
|
|
|
|
|
chrX_+_102318792
|
6.166
|
|
Magee1
|
melanoma antigen, family E, 1
|
|
chr6_-_60779848
|
6.160
|
NM_009221
|
Snca
|
synuclein, alpha
|
|
chr8_+_56040445
|
6.042
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr15_+_3220999
|
5.978
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr18_-_25637339
|
5.924
|
|
Celf4
|
CUGBP, Elav-like family member 4
|
|
chr9_+_111277323
|
5.918
|
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr9_-_49607164
|
5.917
|
NM_001081445
NM_001113204
NM_010875
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr2_+_135883662
|
5.821
|
NM_029530
|
6330527O06Rik
|
RIKEN cDNA 6330527O06 gene
|
|
chr2_+_177876710
|
5.764
|
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr15_-_79656745
|
5.710
|
|
Cbx6
|
chromobox homolog 6
|
|
chr15_+_3220994
|
5.670
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr7_-_149838254
|
5.570
|
|
Igf2
|
insulin-like growth factor 2
|
|
chr2_-_140485267
|
5.537
|
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr10_+_69487700
|
5.510
|
|
Ank3
|
ankyrin 3, epithelial
|
|
chrX_+_54032858
|
5.490
|
NM_001077362
|
Fhl1
|
four and a half LIM domains 1
|
|
chr18_+_37925233
|
5.444
|
NM_033595
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr15_+_3220766
|
5.430
|
NM_001042613
NM_001042614
NM_009155
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chrX_+_54032914
|
5.412
|
|
Fhl1
|
four and a half LIM domains 1
|
|
chr18_+_37925619
|
5.333
|
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr10_+_34018323
|
5.294
|
|
Tspyl4
|
TSPY-like 4
|
|
chr18_+_23656024
|
5.260
|
|
Dtna
|
dystrobrevin alpha
|
|
chr5_+_104868321
|
5.191
|
|
Spp1
|
secreted phosphoprotein 1
|
|
chr8_+_56040080
|
5.189
|
NM_153581
|
Gpm6a
|
glycoprotein m6a
|
|
chr5_+_89012596
|
5.135
|
NM_027530
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr16_-_36367622
|
5.093
|
NM_001082546
|
BC100530
BC117090
|
cDNA sequence BC100530
cDNA sequence BC1179090
|
|
chr1_-_170085342
|
5.091
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr18_+_37821598
|
4.969
|
NM_033584
|
Pcdhga1
|
protocadherin gamma subfamily A, 1
|
|
chr4_-_63681749
|
4.969
|
NM_011607
|
Tnc
|
tenascin C
|
|
chr15_-_101510717
|
4.928
|
NM_010669
|
Krt6b
|
keratin 6B
|
|
chr5_+_29761203
|
4.913
|
NM_001033457
|
Nom1
|
nucleolar protein with MIF4G domain 1
|
|
chr1_+_12708582
|
4.858
|
NM_172294
|
Sulf1
|
sulfatase 1
|
|
chr15_-_8660729
|
4.770
|
NM_148938
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr2_-_52414072
|
4.735
|
NM_146123
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr15_+_97962651
|
4.723
|
|
|
|
|
chr12_+_25265583
|
4.718
|
NM_145890
|
Grhl1
|
grainyhead-like 1 (Drosophila)
|
|
chr7_+_107423813
|
4.688
|
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr2_+_145068346
|
4.641
|
NM_053195
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr14_+_14286471
|
4.641
|
NM_001163032
|
Synpr
|
synaptoporin
|
|
chr8_+_96696512
|
4.616
|
NM_008630
|
Mt2
|
metallothionein 2
|
|
chr5_-_104542598
|
4.614
|
|
Sparcl1
|
SPARC-like 1
|
|
chr8_-_48374715
|
4.562
|
|
Stox2
|
storkhead box 2
|
|
chr10_+_96944961
|
4.554
|
NM_007833
|
Dcn
|
decorin
|
|
chr11_-_55233366
|
4.525
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr5_-_104542641
|
4.519
|
|
Sparcl1
|
SPARC-like 1
|
|
chr8_-_49760037
|
4.431
|
NM_001145937
NM_011857
|
Odz3
|
odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr1_-_171677653
|
4.411
|
NM_009062
|
Rgs4
|
regulator of G-protein signaling 4
|
|
chr6_+_3954259
|
4.405
|
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr9_+_75192367
|
4.334
|
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr7_-_110976314
|
4.312
|
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr5_-_104542729
|
4.268
|
|
Sparcl1
|
SPARC-like 1
|
|
chr16_+_36277233
|
4.253
|
NM_001082543
|
Stfa1
|
stefin A1
|
|
chr17_+_75784448
|
4.236
|
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr18_-_31476527
|
4.220
|
|
Rit2
|
Ras-like without CAAX 2
|
|
chr3_+_14886253
|
4.188
|
NM_009801
|
Car2
|
carbonic anhydrase 2
|
|
chr7_-_110976499
|
4.172
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr8_-_105308910
|
4.150
|
NM_009866
|
Cdh11
|
cadherin 11
|
|
chr7_-_107805466
|
4.137
|
NM_001163186
NM_001163187
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr10_+_40657980
|
4.131
|
|
Wasf1
|
WASP family 1
|
|
chr4_+_104830022
|
4.123
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr7_-_110976438
|
4.102
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr6_+_3953944
|
4.071
|
NM_025331
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr18_-_25637786
|
4.004
|
|
Celf4
|
CUGBP, Elav-like family member 4
|
|
chr10_+_97028134
|
4.003
|
NM_008524
|
Lum
|
lumican
|
|
chr15_-_98783909
|
3.988
|
NM_011653
|
Tuba1a
|
tubulin, alpha 1A
|
|
chr11_-_102331042
|
3.986
|
NM_010575
|
Itga2b
|
integrin alpha 2b
|
|
chr6_+_3954225
|
3.979
|
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr13_+_49704471
|
3.948
|
|
Ogn
|
osteoglycin
|
|
chr11_-_11832366
|
3.944
|
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr9_+_40590943
|
3.864
|
|
9030425E11Rik
|
RIKEN cDNA 9030425E11 gene
|
|
chr16_+_35002066
|
3.827
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr13_+_49703402
|
3.816
|
NM_008760
|
Ogn
|
osteoglycin
|
|
chr14_+_76362274
|
3.809
|
|
Kctd4
|
potassium channel tetramerisation domain containing 4
|
|
chr7_-_107805365
|
3.808
|
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr14_+_28435613
|
3.795
|
NM_177814
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr6_-_121423633
|
3.794
|
|
Iqsec3
|
IQ motif and Sec7 domain 3
|
|
chr10_+_97028409
|
3.788
|
|
Lum
|
lumican
|
|
chr11_+_44431601
|
3.781
|
NM_007897
|
Ebf1
|
early B-cell factor 1
|
|
chr6_-_92517359
|
3.776
|
NM_001134461
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr10_+_98453513
|
3.767
|
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr4_-_138419379
|
3.765
|
NM_001122954
|
Pla2g5
|
phospholipase A2, group V
|
|
chr14_-_20796411
|
3.752
|
NM_001038637
NM_010315
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr18_-_31476721
|
3.739
|
NM_009065
|
Rit2
|
Ras-like without CAAX 2
|
|
chr10_+_97028456
|
3.723
|
|
Lum
|
lumican
|
|
chr1_+_17135421
|
3.716
|
NM_010267
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1
|
|
chr10_-_70055559
|
3.711
|
NM_001162846
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like
|
|
chrX_-_149471740
|
3.711
|
NM_023788
|
Mageh1
|
melanoma antigen, family H, 1
|
|
chrX_-_132749976
|
3.627
|
NM_009052
|
Bex1
|
brain expressed gene 1
|
|
chr19_-_5799376
|
3.598
|
|
Malat1
|
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA)
|
|
chr7_-_110976439
|
3.598
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr7_-_133939596
|
3.594
|
|
|
|
|
chr7_-_147268150
|
3.579
|
NM_001190386
|
Caly
|
calcyon neuron-specific vesicular protein
|
|
chr1_+_155756310
|
3.564
|
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase)
|
|
chr12_+_90191833
|
3.513
|
NM_172544
|
Nrxn3
|
neurexin III
|
|
chr8_-_67071497
|
3.484
|
|
|
|
|
chrX_+_133357268
|
3.484
|
NM_011123
|
Plp1
|
proteolipid protein (myelin) 1
|
|
chr4_+_114761312
|
3.475
|
NM_001164557
NM_001164558
NM_026018
|
Pdzk1ip1
|
PDZK1 interacting protein 1
|
|
chr11_+_87391881
|
3.468
|
|
Sept4
|
septin 4
|
|
chr11_-_30839546
|
3.463
|
|
Erlec1
|
endoplasmic reticulum lectin 1
|
|
chr7_-_147268395
|
3.452
|
NM_001190385
NM_026769
|
Caly
|
calcyon neuron-specific vesicular protein
|
|
chr1_-_45559866
|
3.441
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr9_+_7558476
|
3.438
|
|
Mmp8
|
matrix metallopeptidase 8
|
|
chr8_-_48374755
|
3.419
|
NM_001114311
|
Stox2
|
storkhead box 2
|
|
chr9_+_7558428
|
3.411
|
NM_008611
|
Mmp8
|
matrix metallopeptidase 8
|
|
chr1_+_45368382
|
3.397
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr6_+_121586190
|
3.386
|
NM_175628
|
A2m
|
alpha-2-macroglobulin
|
|
chr9_-_54509453
|
3.370
|
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr11_-_55233339
|
3.369
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr6_+_141472906
|
3.365
|
NM_001177772
NM_021471
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1
|
|
chr18_+_80315134
|
3.334
|
|
Pard6g
|
par-6 partitioning defective 6 homolog gamma (C. elegans)
|
|
chr3_-_92674126
|
3.320
|
NM_025621
|
2310050C09Rik
|
RIKEN cDNA 2310050C09 gene
|
|
chr10_+_96945057
|
3.312
|
|
Dcn
|
decorin
|
|
chr4_+_128820458
|
3.310
|
|
Fndc5
|
fibronectin type III domain containing 5
|
|
chr12_+_96930429
|
3.289
|
NM_201518
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2
|
|
chrX_-_16394443
|
3.280
|
NM_172778
|
Maob
|
monoamine oxidase B
|
|
chr6_-_60778740
|
3.278
|
NM_001042451
|
Snca
|
synuclein, alpha
|
|
chr1_+_152166546
|
3.270
|
NM_001159730
NM_024458
|
Pdc
|
phosducin
|
|
chr15_-_54751515
|
3.267
|
NM_001136077
NM_015744
|
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr7_+_4071087
|
3.251
|
NM_001001454
NM_001109765
NM_021324
|
Ttyh1
|
tweety homolog 1 (Drosophila)
|
|
chr5_-_139606339
|
3.229
|
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta
|
|
chr9_-_52484889
|
3.211
|
|
AI593442
|
expressed sequence AI593442
|
|
chr3_-_126646303
|
3.197
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr11_-_103733519
|
3.197
|
|
Nsf
|
N-ethylmaleimide sensitive fusion protein
|
|
chr3_+_136598184
|
3.179
|
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform
|
|
chr6_-_127075853
|
3.166
|
|
Ccnd2
|
cyclin D2
|
|
chr6_+_56828743
|
3.146
|
|
Fkbp9
|
FK506 binding protein 9
|
|
chr17_+_73186891
|
3.145
|
|
Ypel5
|
yippee-like 5 (Drosophila)
|
|
chr4_+_101180625
|
3.131
|
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr16_-_36131169
|
3.129
|
NM_001033239
|
Csta
|
cystatin A
|
|
chr1_+_43787692
|
3.122
|
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene
|
|
chr4_+_114761644
|
3.079
|
|
Pdzk1ip1
|
PDZK1 interacting protein 1
|
|
chr17_+_73186929
|
3.062
|
|
Ypel5
|
yippee-like 5 (Drosophila)
|
|
chr1_+_43787442
|
3.061
|
NM_024283
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene
|
|
chr11_+_67013553
|
3.035
|
NM_030679
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult
|
|
chr19_-_5800268
|
3.024
|
|
|
|
|
chr14_-_52530382
|
3.016
|
|
Ndrg2
|
N-myc downstream regulated gene 2
|
|
chr7_-_54444724
|
3.014
|
NM_205821
|
Mrgpra6
|
MAS-related GPR, member A6
|
|
chrX_-_147241307
|
3.013
|
|
Maged2
|
melanoma antigen, family D, 2
|
|
chr2_-_62411970
|
2.983
|
NM_007986
|
Fap
|
fibroblast activation protein
|
|
chr18_+_82754829
|
2.962
|
|
Mbp
|
myelin basic protein
|
|
chr1_+_155500166
|
2.948
|
NM_026380
|
Rgs8
|
regulator of G-protein signaling 8
|
|
chr11_+_119862650
|
2.942
|
|
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2
|
|
chr13_-_3803547
|
2.942
|
NM_027416
|
Calml3
|
calmodulin-like 3
|
|
chr3_+_13471681
|
2.936
|
|
Ralyl
|
RALY RNA binding protein-like
|
|
chr1_-_42751652
|
2.933
|
|
2610017I09Rik
|
RIKEN cDNA 2610017I09 gene
|
|
chr11_-_30119771
|
2.921
|
NM_175836
|
Spnb2
|
spectrin beta 2
|
|
chr17_+_72131286
|
2.915
|
NM_030179
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr11_-_83880574
|
2.914
|
|
Dusp14
|
dual specificity phosphatase 14
|
|
chr11_+_94812919
|
2.904
|
|
Col1a1
|
collagen, type I, alpha 1
|
|
chr2_+_177876858
|
2.892
|
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr10_+_90292388
|
2.883
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr14_+_13073221
|
2.875
|
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G
|
|
chr9_+_72859850
|
2.857
|
|
Ccpg1
|
cell cycle progression 1
|
|
chr14_-_9142289
|
2.842
|
NM_183187
|
Fam107a
|
family with sequence similarity 107, member A
|
|
chr2_-_140497204
|
2.825
|
NM_001172160
NM_178382
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr18_-_25912453
|
2.815
|
NM_001146292
NM_001146293
NM_001146294
NM_001146295
NM_001174074
NM_133195
|
Celf4
|
CUGBP, Elav-like family member 4
|
|
chr1_-_45559700
|
2.807
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr17_+_75578120
|
2.776
|
NM_206958
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr14_+_55555300
|
2.772
|
NM_026066
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5
|
|
chrX_-_132738383
|
2.735
|
NM_177919
|
Tceal5
|
transcription elongation factor A (SII)-like 5
|
|
chr12_+_89961317
|
2.690
|
NM_001198587
|
Nrxn3
|
neurexin III
|
|
chr18_+_37644674
|
2.681
|
NM_053142
|
Pcdhb17
|
protocadherin beta 17
|
|
chr7_+_4071171
|
2.681
|
|
Ttyh1
|
tweety homolog 1 (Drosophila)
|
|
chr9_-_54509658
|
2.680
|
NM_053178
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr7_-_88851823
|
2.671
|
|
Homer2
|
homer homolog 2 (Drosophila)
|
|
chr1_+_24184448
|
2.660
|
NM_007740
|
Col9a1
|
collagen, type IX, alpha 1
|
|
chr18_-_43552694
|
2.657
|
NM_009468
|
Dpysl3
|
dihydropyrimidinase-like 3
|
|
chr4_+_134024234
|
2.652
|
NM_019641
|
Stmn1
|
stathmin 1
|
|
chr14_-_56143027
|
2.652
|
NM_008736
|
Nrl
|
neural retina leucine zipper gene
|
|
chr11_+_98629709
|
2.647
|
|
|
|
|
chr8_-_70342105
|
2.643
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|